4GMY
 
 | |
8WGT
 
 | | Crystal structure of V30M-TTR in complex with compound 7 | | Descriptor: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGS
 
 | | Crystal structure of V30M-TTR in complex with compound 4 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-(2-ethyl-4-iodanyl-1-benzofuran-3-yl)methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGU
 
 | | Crystal structure of V30M-TTR in complex with compound 20 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-[2-ethyl-4,7-bis(fluoranyl)-1-benzofuran-3-yl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
4PPH
 
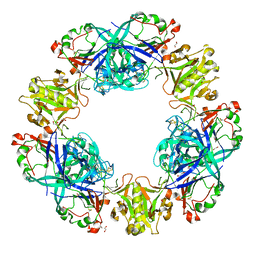 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4I4F
 
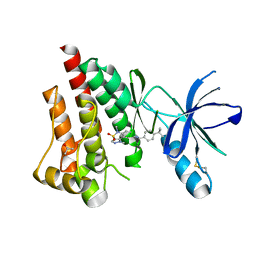 | | Structure of Focal Adhesion Kinase catalytic domain in complex with an allosteric binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, ISOPROPYL ALCOHOL, N-(4-tert-butylbenzyl)-1,5-dimethyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-amine 4,4-dioxide | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5AWG
 
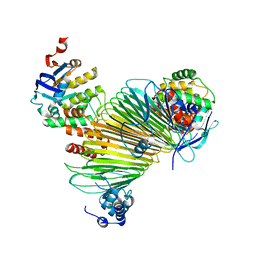 | | Crystal structure of Hg-bound SufB-SufC-SufD complex from Escherichia coli | | Descriptor: | FeS cluster assembly protein SufB, FeS cluster assembly protein SufD, MERCURY (II) ION, ... | | Authors: | Hirabayashi, K, Wada, K. | | Deposit date: | 2015-07-03 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (4.278 Å) | | Cite: | Functional Dynamics Revealed by the Structure of the SufBCD Complex, a Novel ATP-binding Cassette (ABC) Protein That Serves as a Scaffold for Iron-Sulfur Cluster Biogenesis
J.Biol.Chem., 290, 2015
|
|
4I4E
 
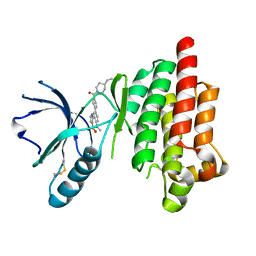 | | Structure of Focal Adhesion Kinase catalytic domain in complex with hinge binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, [4-(2-hydroxyethyl)piperidin-1-yl][4-(5-methyl-4,4-dioxido-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-yl)phenyl]methanone | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1IX4
 
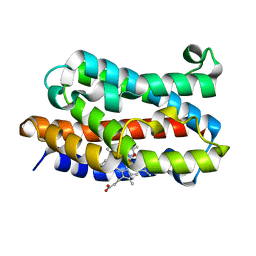 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Carbon Monoxide | | Descriptor: | CARBON MONOXIDE, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1ARP
 
 | |
1IX3
 
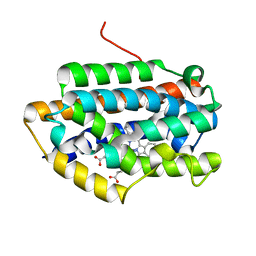 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Cyanide | | Descriptor: | CYANIDE ION, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1EHG
 
 | |
1J10
 
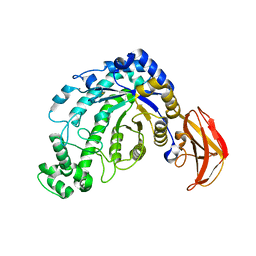 | | beta-amylase from Bacillus cereus var. mycoides in complex with GGX | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J11
 
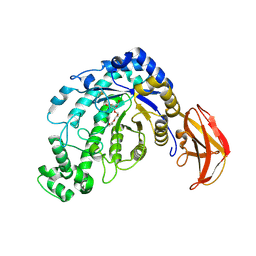 | | beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1IDZ
 
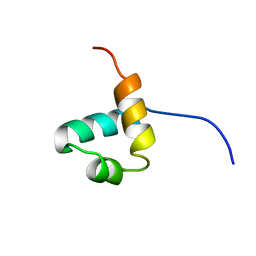 | |
8IHL
 
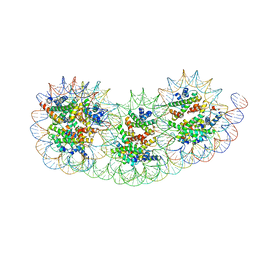 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
1J0Z
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with maltose | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Y
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with glucose | | Descriptor: | Beta-amylase, CALCIUM ION, beta-D-glucopyranose | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1IDY
 
 | |
1UDM
 
 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
6C4D
 
 | | Structure based design of RIP1 kinase inhibitors | | Descriptor: | (3S)-3-(2-benzyl-3-chloro-7-oxo-2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepine-8-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
6C3E
 
 | | CRYSTAL STRUCTURE OF RIP1 KINASE BOUND TO INHIBITOR | | Descriptor: | 2-benzyl-5-nitro-1H-benzimidazole, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
1V6F
 
 | | Solution Structure of Glia Maturation Factor-beta from Mus Musculus | | Descriptor: | glia maturation factor, beta | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-05-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
4GJ3
 
 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4GII
 
 | |
