4MZ0
 
 | |
4NSI
 
 | | Carboplatin binding to HEWL in 20% propanol, 20% PEG 4000 at pH5.6 | | Descriptor: | CITRATE ANION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4MGS
 
 | | BiXyn10A CBM1 APO | | Descriptor: | Putative glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4NSJ
 
 | | Carboplatin binding to HEWL in 2M NH4formate, 0.1M HEPES at pH 7.5 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4O2Z
 
 | | Crystal Structure of MPK3 from Leishmania donovani, LdBPK_100540 in the presence of NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, Mitogen-activated protein kinase 3, putative | | Authors: | Wernimont, A.K, Walker, J.R, Hutchinson, A, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Hui, R, Mangos, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of MPK3 from Leishmania donovani, LdBPK_100540 in the presence of NVP-BBT594
To be Published
|
|
4MYY
 
 | |
4MYZ
 
 | |
4MT1
 
 | |
4N4P
 
 | | Crystal Structure of N-acetylneuraminate lyase from Mycoplasma synoviae, crystal form I | | Descriptor: | Acylneuraminate lyase, CHLORIDE ION | | Authors: | Georgescauld, F, Popova, K, Gupta, A.J, Bracher, A, Engen, J.R, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2013-10-08 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding.
Cell(Cambridge,Mass.), 157, 2014
|
|
4NU6
 
 | | Crystal Structure of PTDH R301K | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4O2W
 
 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ODH
 
 | | Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.UCA heavy chain, CAP256-VRC26.UCA light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OCS
 
 | | Crystal structure of human Fab CAP256-VRC26.10, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.10 heavy chain, CAP256-VRC26.10 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4LT1
 
 | | HEWL co-crystallised with Carboplatin in non-NaCl conditions: crystal 1 processed using the XDS software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
4OWA
 
 | |
4OWE
 
 | | PtCl6 binding to HEWL | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4OD3
 
 | | Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CAP256-VRC26.07 heavy chain, CAP256-VRC26.07 light chain, ... | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4MAB
 
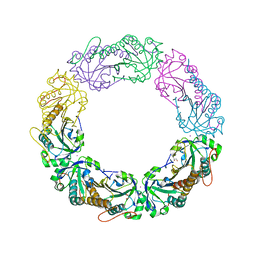 | | Resolving Cys to Ala variant of Salmonella Alkyl Hydroperoxide Reductase C in its substrate-ready conformation | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, GLYCEROL, ... | | Authors: | Perkins, A, Nelson, K.J, Williams, J.R, Poole, L.B, Karplus, P.A. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sensitive balance between the fully folded and locally unfolded conformations of a model peroxiredoxin.
Biochemistry, 52, 2013
|
|
4OQJ
 
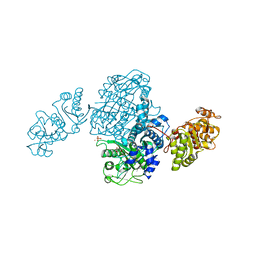 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ORG
 
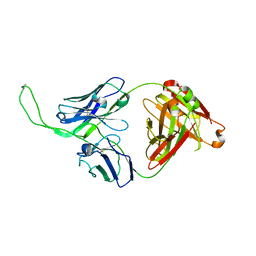 | | Crystal structure of human Fab CAP256-VRC26.04, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.04 heavy chain, CAP256-VRC26.04 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.121 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OH3
 
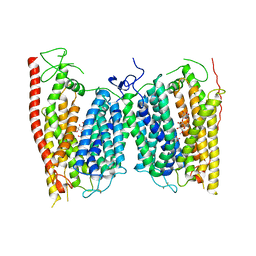 | | Crystal structure of a nitrate transporter | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, NITRATE ION, Nitrate transporter 1.1 | | Authors: | Sun, J, Bankston, J.R, Payandeh, J, Hinds, T.R, Zagotta, W.N, Zheng, N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-05 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the plant dual-affinity nitrate transporter NRT1.1.
Nature, 507, 2014
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
