7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F39
 
 | | The structure of flavin transferase FmnB | | Descriptor: | FAD:protein FMN transferase | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F2U
 
 | | FmnB complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-14 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
6KXA
 
 | | Galectin-3 CRD binds to GalA dimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
7VVB
 
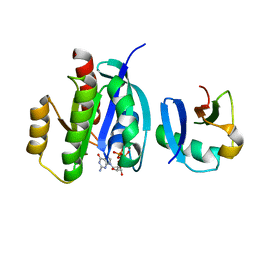 | |
7VV9
 
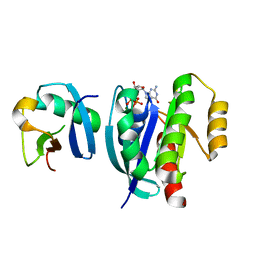 | |
7VVG
 
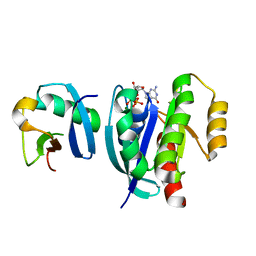 | |
7VV8
 
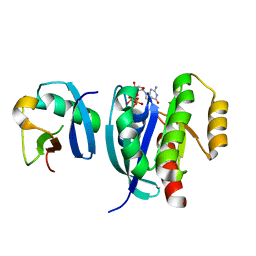 | |
5O71
 
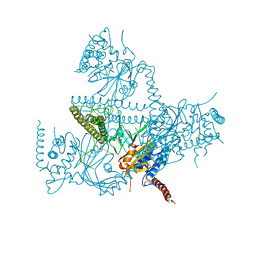 | | Crystal structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Reverter, D, Liu, B. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25.
Nat Commun, 9, 2018
|
|
8XQ9
 
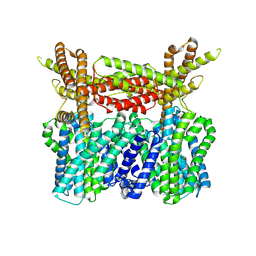 | |
8XQA
 
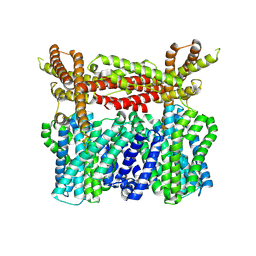 | |
8XQ4
 
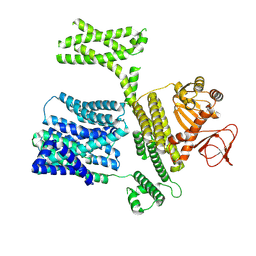 | |
8XPQ
 
 | |
8XQ7
 
 | |
8XQ8
 
 | |
9IJ1
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, bilobed) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIY
 
 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, bilobed) | | Descriptor: | Piwi, RNA (5'-R(P*CP*GP*UP*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*GP*AP*UP*CP*AP*GP*CP*U)-3'), RNA (5'-R(P*UP*AP*GP*CP*AP*GP*AP*UP*CP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*AP*CP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ4
 
 | | Cryo-EM Structure of MILI(K852A)-piRNA-target (26-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ5
 
 | | Cryo-EM Structure of MILI(K853A)-piRNA-target | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ3
 
 | | Cryo-EM Structure of MILI-piRNA-target (26-nt) | | Descriptor: | MANGANESE (II) ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ0
 
 | | Cryo-EM Structure of MILI-piRNA-target (8-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ2
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, comma) | | Descriptor: | Piwi-like protein 2, RNA (5'-R(P*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3'), RNA (5'-R(P*UP*UP*AP*CP*CP*AP*UP*CP*AP*AP*CP*AP*UP*GP*GP*AP*AP*AP*CP*UP*UP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIZ
 
 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, comma) | | Descriptor: | Piwi, RNA (25-MER), RNA (5'-R(P*AP*GP*CP*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
7V1N
 
 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
