8W4G
 
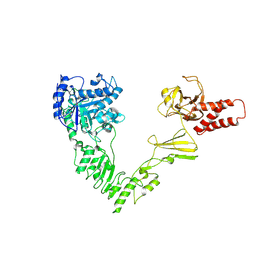 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4I
 
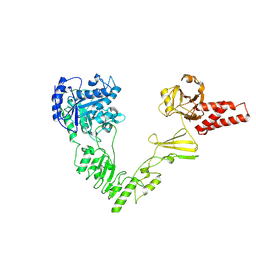 | | Crystal structure of EndoSz mutant D234M in space group P21 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4N
 
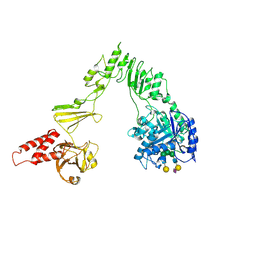 | | Crystal structure of EndoSz mutant D234M, in space group P21, in complex with oligosaccharide G2S1 | | Descriptor: | CALCIUM ION, Glycoside hydrolase, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
6AB6
 
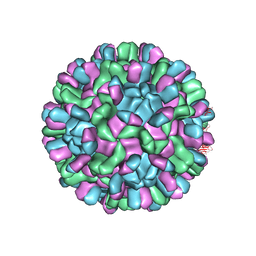 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
2LU1
 
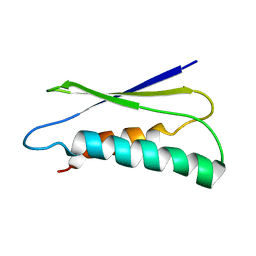 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
5XMJ
 
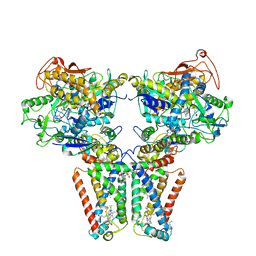 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
6AB5
 
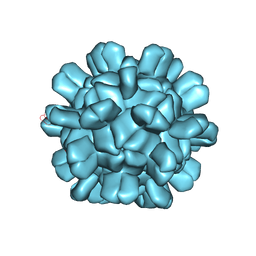 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
1SUE
 
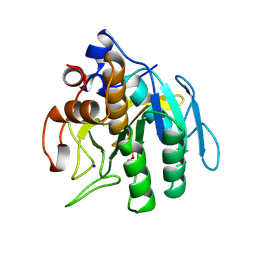 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
4RFU
 
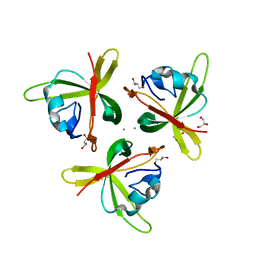 | | Crystal structure of truncated P-domain from Grouper nervous necrosis virus capsid protein at 1.2A | | Descriptor: | CALCIUM ION, Coat protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4RFT
 
 | | T=1 subviral particle of Grouper nervous necrosis virus capsid protein deletion mutant (delta 1-34 & 218-338) | | Descriptor: | Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
2MH8
 
 | | GA-79-MBP cs-rosetta structures | | Descriptor: | GA-79-MBP, maltose binding protein | | Authors: | He, Y, Chen, Y, Porter, L, Bryan, P, Orban, J. | | Deposit date: | 2013-11-19 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Subdomain interactions foster the design of two protein pairs with 80% sequence identity but different folds.
Biophys.J., 108, 2015
|
|
3AM3
 
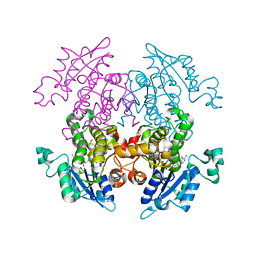 | | A372M mutant of Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan | | Descriptor: | Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Maity, K, Banerjee, T, Narayanappa, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of substrate binding loop mutations on the structure, kinetics, and inhibition of enoyl acyl carrier protein reductase from plasmodium falciparum
Iubmb Life, 63, 2011
|
|
3AM4
 
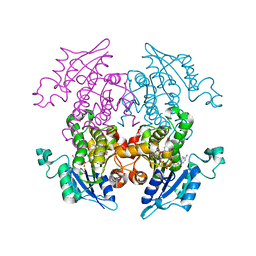 | | A372M mutant of Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T1 | | Descriptor: | 4-(2,4-dichlorophenoxy)-3-hydroxybenzaldehyde, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Maity, K, Banerjee, T, Narayanappa, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of substrate binding loop mutations on the structure, kinetics, and inhibition of enoyl acyl carrier protein reductase from plasmodium falciparum
Iubmb Life, 63, 2011
|
|
3AM5
 
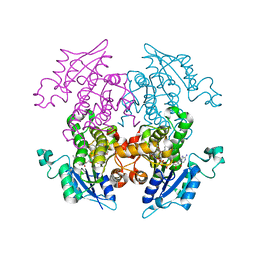 | | K316A mutant of Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan | | Descriptor: | Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Maity, K, Banerjee, T, Narayanappa, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effect of substrate binding loop mutations on the structure, kinetics, and inhibition of enoyl acyl carrier protein reductase from plasmodium falciparum
Iubmb Life, 63, 2011
|
|
