7CZW
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D00
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-1B8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,chain H of FabP5A-1B8,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D03
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
5YVE
 
 | |
6JFI
 
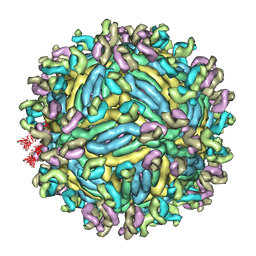 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
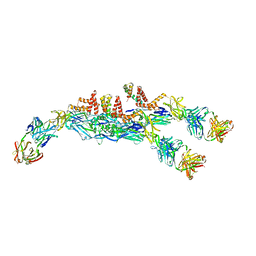 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JWE
 
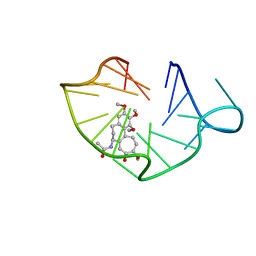 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
9IQX
 
 | | Cryo-EM structure of the human TRPV4-RhoA in complex with AH001 | | Descriptor: | (1~{R})-1-(3-ethylphenyl)ethane-1,2-diol, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Ruan, S.S, Li, S.L. | | Deposit date: | 2024-07-13 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Inactivation of RhoA for Hypertension Treatment Through the TRPV4-RhoA-RhoGDI1 Axis.
Circulation, 2025
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9J3M
 
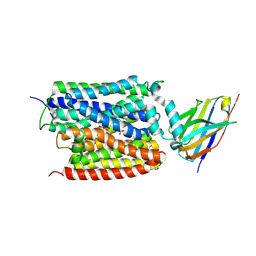 | | ADP/Pi bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3L
 
 | | ATP bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3N
 
 | | ATP bound Chlamydia pneumoniae ATP/ADP translocator NTT1(Inward open state) | | Descriptor: | 1D10, ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3O
 
 | | Chlamydia pneumoniae ATP/ADP translocator NTT1(Outward open state) | | Descriptor: | 1D10, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3J
 
 | | Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADP,ATP carrier protein 1, chloroplastic, nanobody: B-C8 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
8ZTP
 
 | | Crystal structure of cysteine desulfurase Sufs from Mycoplasma Pneumonia | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, cysteine desulfurase | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
8ZTQ
 
 | | Crystal structure of Sufu from Mycoplasma Pneumonia | | Descriptor: | Nitrogen fixation protein NifU, ZINC ION | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
8X82
 
 | |
8X83
 
 | |
8X84
 
 | |
6LI6
 
 | | Crystal structure of MCR-1-S treated by Au(PEt3)Cl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1, TRIETHYLPHOSPHANE | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
8IDM
 
 | | Crystal structure of nanobody VHH-227 with nanobody VHH-T71 and MERS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IDO
 
 | | Crystal structure of nanobody VHH-T148 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T148, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IFN
 
 | | MERS-CoV spike trimer in complex with nanobody VHH-T148 | | Descriptor: | Spike glycoprotein, VHH-T148, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
