7CYV
 
 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
7XEF
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with (R)-(1-benzyl-3-phenylpyrrolidin-3-yl)methanol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XEE
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with 2-(3-phenyloxetan-3-yl)ethanamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-phenyloxetan-3-yl)ethanamine, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
7VWD
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VX2
 
 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWM
 
 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-11 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7YF0
 
 | | In situ structure of polymerase complex of mammalian reovirus in the core | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YED
 
 | | In situ structure of polymerase complex of mammalian reovirus in the elongation state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Lambda-2 protein, MAGNESIUM ION, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YEZ
 
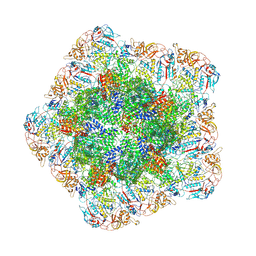 | | In situ structure of polymerase complex of mammalian reovirus in the reloaded state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFE
 
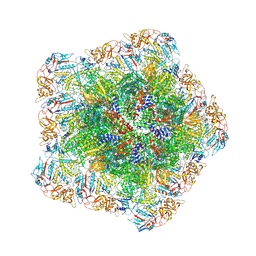 | | In situ structure of polymerase complex of mammalian reovirus in virion | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA (5'-R(P*AP*CP*GP*AP*UP*UP*AP*GP*C)-3'), ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YEV
 
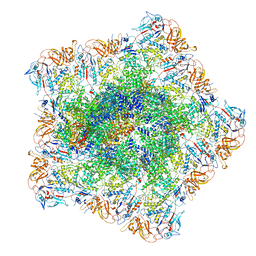 | | In situ structure of polymerase complex of mammalian reovirus in the pre-elongation state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XY9
 
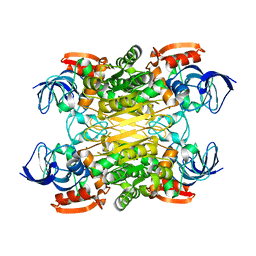 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
7EDO
 
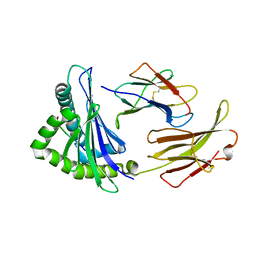 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
7WZA
 
 | | An open conformation Form 1 of switch II for RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50028777 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
7WZC
 
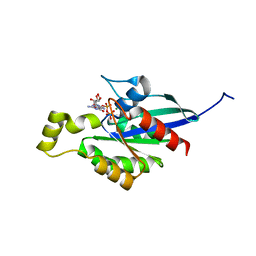 | | An open conformation Form2 of switch II for RhoA GDP-bound state | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79944921 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
5YFB
 
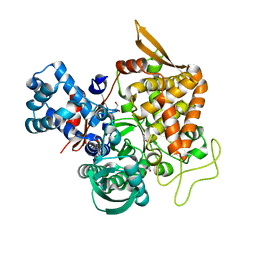 | | Crystal structure of a new DPP III family member | | Descriptor: | 1,2-ETHANEDIOL, Dipeptidyl peptidase 3, GLYCEROL, ... | | Authors: | Xu, T, Sun, Z, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
7C1E
 
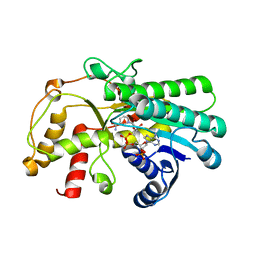 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Y127W) | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, Y.F, Zhou, J.Y, Liu, Y.F, Xu, G.C, Ni, Y. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering an Alcohol Dehydrogenase from Kluyveromyces polyspora for Efficient Synthesis of Ibrutinib Intermediate
Adv.Synth.Catal., 2021
|
|
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Chen, R.C, Gao, S.S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
7WNW
 
 | |
7WNN
 
 | |
7XE8
 
 | |
