2KNV
 
 | |
2MZX
 
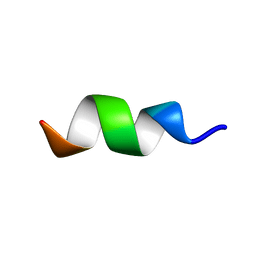 | | CCR5-ECL2 helical structure, residues Q186-T195 | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Abayev, M, Anglister, J. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An extended CCR5 ECL2 peptide forms a helix that binds HIV-1 gp120 through non-specific hydrophobic interactions.
Febs J., 282, 2015
|
|
6FTB
 
 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | Authors: | Punekar, A.S, Dowson, C.J, Roper, D.I. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
1HBR
 
 | | R-STATE FORM OF CHICKEN HEMOGLOBIN D | | Descriptor: | PROTEIN (HEMOGLOBIN D), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, E, Oliveira, M.A, Xie, Q, Ernst, S.R, Riggs, A.F, Hackert, M.L. | | Deposit date: | 1998-12-06 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural and functional analysis of the hemoglobin D component from chicken.
J.Biol.Chem., 274, 1999
|
|
1HBB
 
 | |
1HBA
 
 | |
1RVD
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3BHI
 
 | |
1BZ0
 
 | | HEMOGLOBIN A (HUMAN, DEOXY, HIGH SALT) | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1998-11-04 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accommodation of insertions in helices: the mutation in hemoglobin Catonsville (Pro 37 alpha-Glu-Thr 38 alpha) generates a 3(10)-->alpha bulge.
Biochemistry, 32, 1993
|
|
3D5Q
 
 | | Crystal Structure of 11b-HSD1 in Complex with Triazole Inhibitor | | Descriptor: | 3-[1-(4-fluorophenyl)cyclopropyl]-4-(1-methylethyl)-5-[4-(trifluoromethoxy)phenyl]-4H-1,2,4-triazole, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P.C. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Distinctive molecular inhibition mechanisms for selective inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1.
Bioorg.Med.Chem., 16, 2008
|
|
1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3BHJ
 
 | | Crystal structure of human Carbonyl Reductase 1 in complex with glutathione | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | Authors: | Rauh, D, Bateman, R.L, Shokat, K.M. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
3BHM
 
 | | Crystal structure of human Carbonyl Reductase 1 in complex with S-hydroxymethylglutathione | | Descriptor: | 2-AMINO-4-[1-CARBOXYMETHYL-CARBAMOYL)-2-HYDROXYMETHYLSULFANYL-ETHYLCARBAMOYL]-BUTYRIC ACID, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | Authors: | Bateman, R.L, Rauh, D, Shokat, K.M. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
7W1R
 
 | | Crystal structure of human Suv3 monomer | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial | | Authors: | Jain, M, Yuan, H.S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimeric assembly of human Suv3 helicase promotes its RNA unwinding function in mitochondrial RNA degradosome for RNA decay.
Protein Sci., 31, 2022
|
|
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2PMW
 
 | | The Crystal Structure of Proprotein convertase subtilisin kexin type 9 (PCSK9) | | Descriptor: | Proprotein convertase subtilisin/kexin type 9, SULFATE ION | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of PCSK9: A Regulator of Plasma LDL-Cholesterol.
Structure, 15, 2007
|
|
4MCN
 
 | | Human SOD1 C57S Mutant, Metal-free | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sea, K, Sohn, S.H, Durazo, A, Sheng, Y, Shaw, B, Cao, X, Taylor, A.B, Whitson, L.J, Holloway, S.P, Hart, P.J, Cabelli, D.E, Gralla, E.B, Valentine, J.S. | | Deposit date: | 2013-08-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase.
J.Biol.Chem., 290, 2015
|
|
5U69
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U8A
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U8F
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U6D
 
 | |
4HHB
 
 | |
7KFU
 
 | | Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KFT
 
 | | Partial Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KKZ
 
 | | Crystal structure of mouse anti-HIV potent neutralizing antibody M4H2K1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Heavy chain of Fab fragment of mouse monoclonal antibody M4H2K1, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Neutralizing Antibodies Induced by First-Generation gp41-Stabilized HIV-1 Envelope Trimers and Nanoparticles.
Mbio, 12, 2021
|
|
