7WFG
 
 | | Subcomplexes A and E in NDH complex from Arabidopsis | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase subunit H, chloroplastic, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
8JL1
 
 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W4A
 
 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-08-23 | | Release date: | 2024-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7XRW
 
 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
1QK7
 
 | |
1ZDM
 
 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
7C8D
 
 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
8HTU
 
 | | Cryo-EM structure of PpPSI-L | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Li, M, Pan, X.W, Sun, H.Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
7D0J
 
 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
8GU7
 
 | |
7XZ9
 
 | |
7F9O
 
 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7WKX
 
 | | IL-17A in complex with the humanized antibody HB0017 | | Descriptor: | ACETIC ACID, Heavy chain of HB0017 Fab, Interleukin-17A, ... | | Authors: | Xu, J, Zhu, X, He, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural and functional insights into a novel pre-clinical-stage antibody targeting IL-17A for treatment of autoimmune diseases.
Int.J.Biol.Macromol., 202, 2022
|
|
8I5B
 
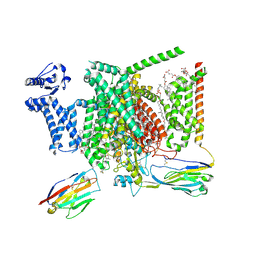 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
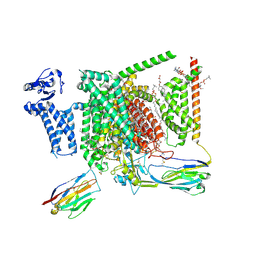 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5G
 
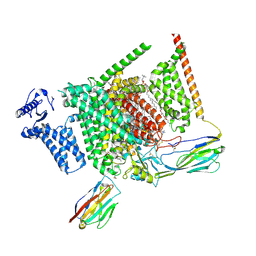 | | Structure of human Nav1.7 in complex with PF-05089771 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8J4F
 
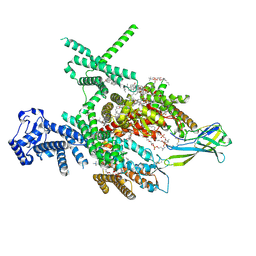 | | Structure of human Nav1.7 in complex with Hardwickii acid | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
7XBG
 
 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFX
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HG0
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFZ
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
