3NUA
 
 | | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens
To be Published
|
|
3R23
 
 | | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, D-alanine--D-alanine ligase | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-12 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis
To be Published
|
|
3QU1
 
 | | Peptide deformylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Peptide deformylase 2, SULFATE ION, ... | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide deformylase from Vibrio cholerae.
To be Published
|
|
3QZ3
 
 | | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Ferritin | | Authors: | Tan, K, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3IIE
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-ray crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis.
To be Published
|
|
3BIO
 
 | | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83.
To be Published
|
|
3B8B
 
 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
3C2Q
 
 | | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, Uncharacterized conserved protein | | Authors: | Duke, N, Gu, M, Mulligan, R, Conrad, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2
To be Published
|
|
1Q4B
 
 | |
5U63
 
 | | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Michalska, K, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae
To Be Published
|
|
1Q4E
 
 | |
1Q4A
 
 | |
1Q4D
 
 | |
1Q4C
 
 | |
6YV7
 
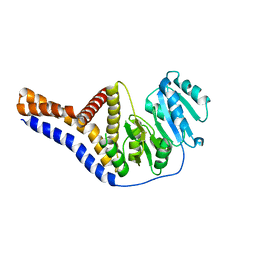 | | Mannosyltransferase PcManGT from Pyrobaculum calidifontis | | Descriptor: | Glycosyl transferase, family 2 | | Authors: | Divne, C, Rosaria, G. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Transmembrane Crenarchaeal Mannosyltransferase Is Involved in N-Glycan Biosynthesis and Displays an Unexpected Minimal Cellulose-Synthase-like Fold.
J.Mol.Biol., 432, 2020
|
|
9FGP
 
 | |
3BM3
 
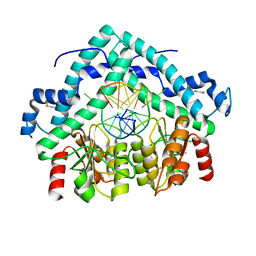 | | Restriction endonuclease PspGI-substrate DNA complex | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3'), ... | | Authors: | Szczepanowski, R.H, Carpenter, M, Czapinska, H, Tamulaitis, G, Siksnys, V, Bhagwat, A, Bochtler, M. | | Deposit date: | 2007-12-12 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A direct crystallographic demonstration that Type II restriction endonuclease PspGI flips nucleotides
To be Published
|
|
5JBQ
 
 | |
2A02
 
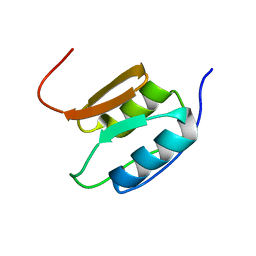 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
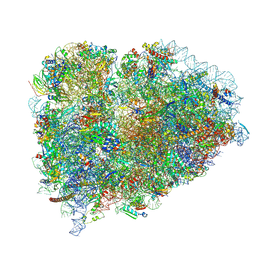
9BDP
 
 | |
9BDL
 
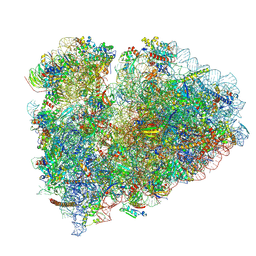 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
6VIN
 
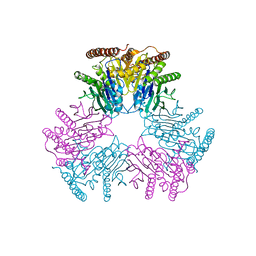 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
5BK3
 
 | | Crystal structure of the neutralizing anti-circumsporozoite protein 580 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 580 Antibody, heavy chain, ... | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
5BK5
 
 | | Crystal structure of the anti-circumsporozoite protein 663 germline antibody | | Descriptor: | 663 germline antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
