5X7K
 
 | |
2K9E
 
 | |
3R8R
 
 | |
3R5E
 
 | |
5NEN
 
 | |
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
1H89
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1JQN
 
 | |
1JQO
 
 | |
4WBK
 
 | | The 1.37 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular Dynamics Simulations of Heart-type Fatty Acid Binding Protein in Apo and Holo Forms, and Hydration Structure Analyses in the Binding Cavity
J.Phys.Chem.B, 119, 2015
|
|
5AX9
 
 | | Crystal structure of the kinase domain of human TRAF2 and NCK-interacting protein kinase in complex with compund 9 | | Descriptor: | 4-methoxy-3-[2-[(3-methoxy-4-morpholin-4-yl-phenyl)amino]pyridin-4-yl]benzenecarbonitrile, SULFATE ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
3E6Y
 
 | | Structure of 14-3-3 in complex with the differentiation-inducing agent Cotylenin A | | Descriptor: | 14-3-3-like protein C, CHLORIDE ION, Cotylenin A, ... | | Authors: | Ottmann, C, Weyand, M, Wittinghofer, A, Oecking, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural rationale for selective stabilization of anti-tumor interactions of 14-3-3 proteins by cotylenin A
J.Mol.Biol., 386, 2009
|
|
4IHL
 
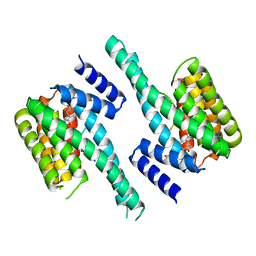 | | Human 14-3-3 isoform zeta in complex with a diphoyphorylated C-RAF peptide and Cotylenin A | | Descriptor: | (1R,3aS,4R,5R,6R,9aR,10E)-6-({(1S,2R,4S,5R,6R,8S,9S)-5-hydroxy-2-(methoxymethyl)-9-methyl-9-[(2S)-oxiran-2-yl]-3,7,10,1 1-tetraoxatricyclo[6.2.1.0~1,6~]undec-4-yl}oxy)-1-(methoxymethyl)-4,9a-dimethyl-7-(propan-2-yl)-1,2,3,3a,4,5,6,8,9,9a-de cahydrodicyclopenta[a,d][8]annulene-1,5-diol, 14-3-3 protein zeta/delta, POTASSIUM ION, ... | | Authors: | Molzan, M, Ottmann, C. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of Physical RAF/14-3-3 Interaction by Cotylenin A as Treatment Strategy for RAS Mutant Cancers.
Acs Chem.Biol., 8, 2013
|
|
4IEA
 
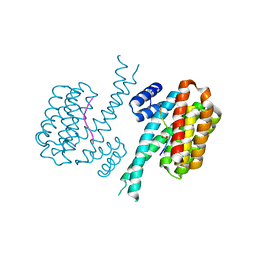 | |
1H88
 
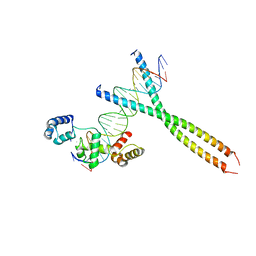 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1IO4
 
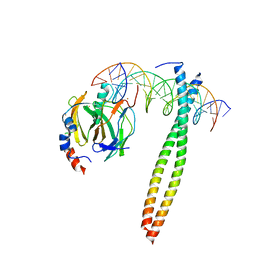 | |
2MP8
 
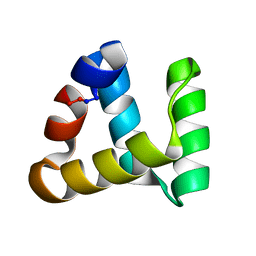 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
6DSQ
 
 | |
6DSS
 
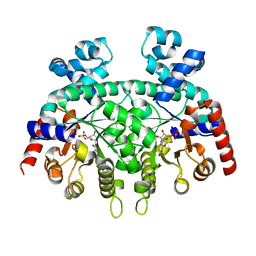 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DSR
 
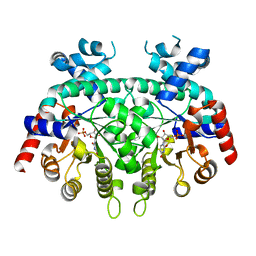 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5D7A
 
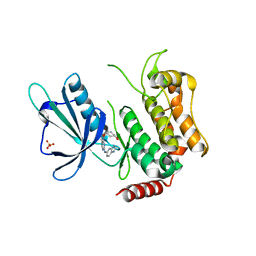 | | Crystal structure of the kinase domain of TRAF2 and NCK-interacting protein kinase with NCB-0846 | | Descriptor: | SULFATE ION, TRAF2 and NCK-interacting protein kinase, cis-4-{[2-(1H-benzimidazol-5-ylamino)quinazolin-8-yl]oxy}cyclohexanol | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
5CWZ
 
 | |
