5FHM
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-(aminomethyl)benzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at resolution 1.55 A resolution | | Descriptor: | (2~{S})-3-[5-[2-[[3-(aminomethyl)phenyl]methyl]-1,2,3,4-tetrazol-5-yl]-3-oxidanyl-1,2-oxazol-4-yl]-2-azanyl-propanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Al-musaed, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
5FHO
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-chlorobenzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at 2.3 A resolution | | Descriptor: | (1S)-1-carboxy-2-(5-{2-[(3-chlorophenyl)methyl]-2H-tetrazol-5-yl}-3-oxo-2,3-dihydro-1,2-oxazol-4-yl)ethan-1-aminium, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
5FHN
 
 | |
6KOF
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 47 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]thiourea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KW7
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 12 | | Descriptor: | 3-(4-bromophenyl)imidazo[2,1-b][1,3]thiazole, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-09-06 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
4JQ7
 
 | | Crystal structure of EGFR kinase domain in complex with compound 2a | | Descriptor: | (2S)-2-[(5,6-diphenylfuro[2,3-d]pyrimidin-4-yl)amino]-2-phenylethanol, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JRV
 
 | | Crystal structure of EGFR kinase domain in complex with compound 4c | | Descriptor: | 4-(dimethylamino)-N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]butanamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
7YLE
 
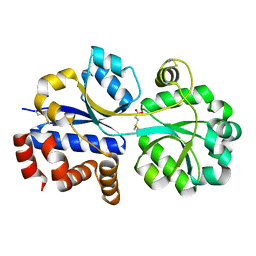 | | RnDmpX in complex with DMSP | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, Glycine betaine/proline ABC transporter, periplasmic glycinebetaine/proline-binding protein | | Authors: | Li, C.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria.
Isme J, 17, 2023
|
|
6GD5
 
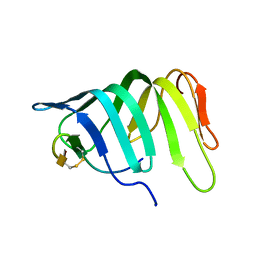 | | The solution structure of the LptA-Thanatin complex | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Moehle, K, Zerbe, O. | | Deposit date: | 2018-04-22 | | Release date: | 2018-11-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport inEscherichia coli.
Sci Adv, 4, 2018
|
|
6KPS
 
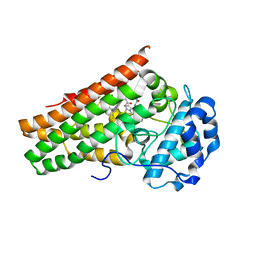 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 36 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]urea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KYC
 
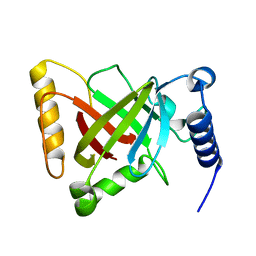 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
6JZ0
 
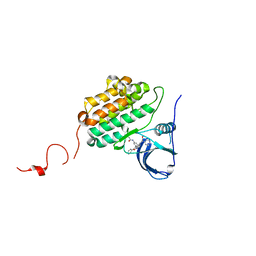 | | Crystal structure of EGFR kinase domain in complex with compound 78 | | Descriptor: | E-4-(dimethylamino)-N-[3-[4-[[(1S)-2-oxidanyl-1-phenyl-ethyl]amino]-6-phenyl-furo[2,3-d]pyrimidin-5-yl]phenyl]but-2-enamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a Furanopyrimidine-Based Epidermal Growth Factor Receptor Inhibitor (DBPR112) as a Clinical Candidate for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 62, 2019
|
|
4JQ8
 
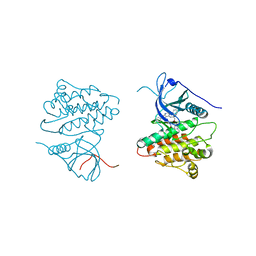 | | Crystal structure of EGFR kinase domain in complex with compound 4b | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]-N~3~,N~3~-dimethyl-beta-alaninamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JR3
 
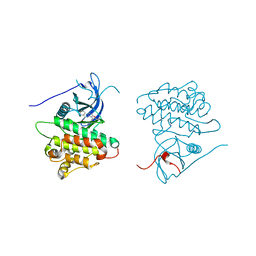 | | Crystal structure of EGFR kinase domain in complex with compound 3g | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]acetamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
6KYD
 
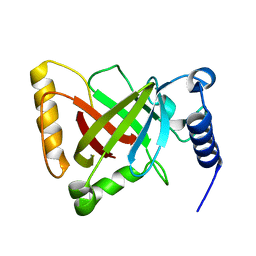 | | Structure of the R217A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
8HXP
 
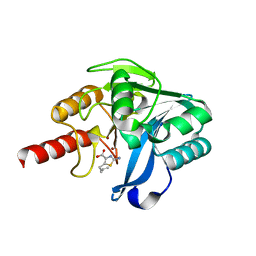 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-(but-3-en-1-yl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-but-3-enyl-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXO
 
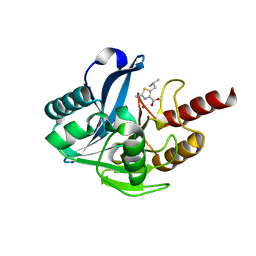 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-isobutylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-methylpropyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, GLYCEROL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXU
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-pentylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-pentyl-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXE
 
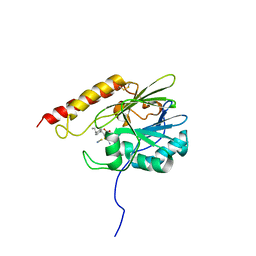 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-propylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Metallo-beta-lactamase L1 type 3, THIOCYANATE ION, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HY2
 
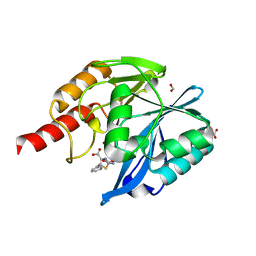 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-phenethylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-phenylethyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXN
 
 | | Crystal structure of B2 Sfh-I MBL in complex with 2-amino-5-(4-(but-3-en-1-yloxy)benzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-but-3-enoxyphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Beta-lactamase, ETHANOL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXW
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-heptylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-heptyl-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HY6
 
 | | Crystal structure of B1 NDM-1 MBL in complex with 2-amino-5-phenethylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-phenylethyl)-1,3-thiazole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXI
 
 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-isopropylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propan-2-ylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase L1 type 3, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HY1
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-(thiophen-2-ylmethyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(thiophen-2-ylmethyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
