5OXI
 
 | | C-terminally retracted ubiquitin L67S mutant | | Descriptor: | SULFATE ION, Ubiquitin L67S mutant | | Authors: | Gladkova, C.G, Schubert, A.F, Wagstaff, J.L, Pruneda, J.N, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5OXH
 
 | | C-terminally retracted ubiquitin T66V/L67N mutant | | Descriptor: | SULFATE ION, Ubiquitin T66V/L67N mutant | | Authors: | Gladkova, C, Schubert, A.F, Wagstaff, J.L, Pruneda, J.P, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
6FPQ
 
 | | Structure of S. pombe Mmi1 in complex with 7-mer RNA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RNA (5'-R(*UP*UP*AP*AP*AP*CP*C)-3'), YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6FPX
 
 | | Structure of S. pombe Mmi1 in complex with 11-mer RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*UP*UP*UP*AP*AP*AP*CP*CP*UP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6FPP
 
 | | Structure of S. pombe Mmi1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5AF4
 
 | | Structure of Lys33-linked diUb | | Descriptor: | GLYCEROL, SULFATE ION, UBIQUITIN | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | Descriptor: | TRABID, UBIQUITIN, ZINC ION | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
4WZP
 
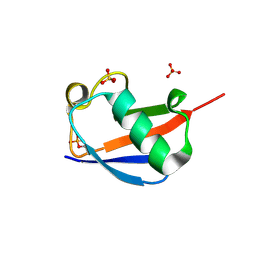 | | Ser65 phosphorylated ubiquitin, major conformation | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Wauer, T, Wagstaff, J, Freund, S.M.V, Komander, D. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis.
Embo J., 34, 2015
|
|
6QIK
 
 | |
6QT0
 
 | |
6QTZ
 
 | |
8QQF
 
 | | TBC1D23 PH domain complexed with STX16 TLY motif | | Descriptor: | GLYCEROL, PHOSPHATE ION, Syntaxin-16, ... | | Authors: | Kaufman, J.G, Owen, D.J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cargo selective vesicle tethering: The structural basis for binding of specific cargo proteins by the Golgi tether component TBC1D23.
Sci Adv, 10, 2024
|
|
6HEM
 
 | |
6HEJ
 
 | | Structure of human USP28 | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEH
 
 | | Structure of the catalytic domain of USP28 (insertion deleted) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28,Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEI
 
 | |
6HEL
 
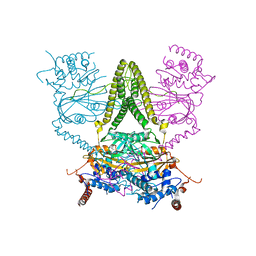 | | Structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEK
 
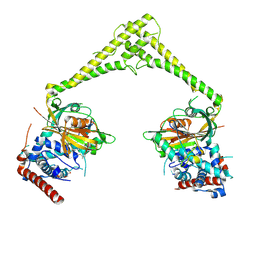 | | Structure of human USP28 bound to Ubiquitin-PA | | Descriptor: | CHLORIDE ION, Polyubiquitin-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6RZZ
 
 | |
6RI5
 
 | |
6S05
 
 | |
8PPO
 
 | |
8Q2J
 
 | | Tau - AD-MIA2 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
