1QHV
 
 | | HUMAN ADENOVIRUS SEROTYPE 2 FIBRE HEAD | | Descriptor: | PROTEIN (ADENOVIRUS FIBRE), SULFATE ION | | Authors: | Van Raaij, M.J, Louis, N, Chroboczek, J, Cusack, S. | | Deposit date: | 1999-05-28 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the human adenovirus serotype 2 fiber head domain at 1.5 A resolution.
Virology, 262, 1999
|
|
1QIU
 
 | |
1COW
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1F5W
 
 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
1EAJ
 
 | |
5LYE
 
 | |
3J4B
 
 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
4UXE
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 selenomethionine crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-19 | | Last modified: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4V0S
 
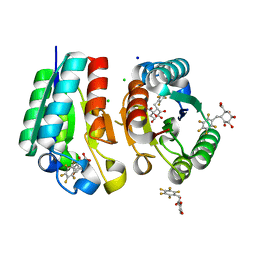 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4UXF
 
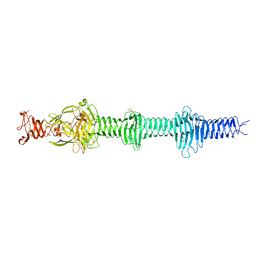 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
7Q4T
 
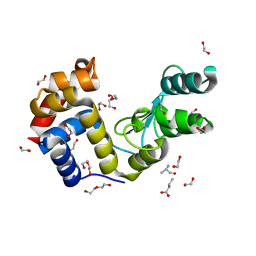 | |
7Q4S
 
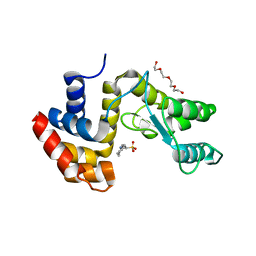 | |
4UMI
 
 | |
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
7QXC
 
 | |
7QXE
 
 | |
7QXD
 
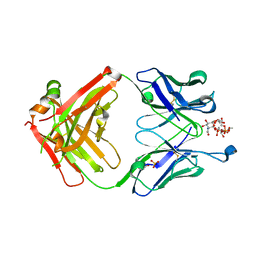 | |
5FX0
 
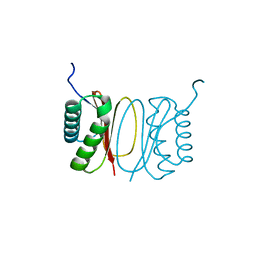 | |
5AQ5
 
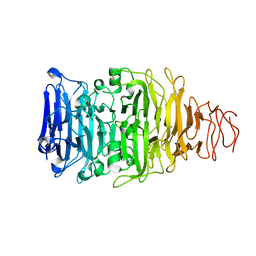 | |
5NXH
 
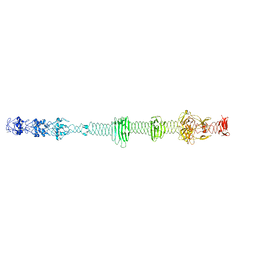 | |
5NXF
 
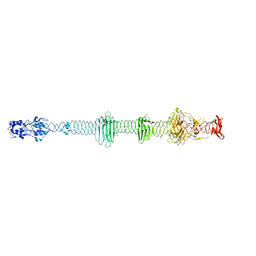 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, residues 795 to 1289, at 1.9 Angstrom. | | Descriptor: | ACETATE ION, GLYCEROL, Long-tail fiber proximal subunit, ... | | Authors: | Namura, M, van Raaij, M.J, Kanamaru, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2A
 
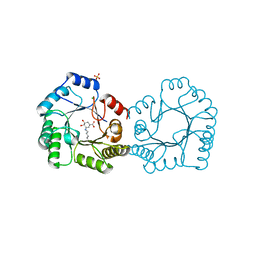 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
