3ICB
 
 | | THE REFINED STRUCTURE OF VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN FROM BOVINE INTESTINE. MOLECULAR DETAILS, ION BINDING, AND IMPLICATIONS FOR THE STRUCTURE OF OTHER CALCIUM-BINDING PROTEINS | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN, SULFATE ION | | Authors: | Szebenyi, D.M.E, Moffat, K. | | Deposit date: | 1986-09-09 | | Release date: | 1986-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins.
J.Biol.Chem., 261, 1986
|
|
1EJI
 
 | | RECOMBINANT SERINE HYDROXYMETHYLTRANSFERASE (MOUSE) | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Szebenyi, D.M.E, Liu, X, Kriksunov, I.A, Stover, P.J, Thiel, D.J. | | Deposit date: | 2000-03-02 | | Release date: | 2000-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers.
Biochemistry, 39, 2000
|
|
1RV4
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1RVU
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1RVY
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1RV3
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | GLYCINE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
5BQ1
 
 | | Capturing Carbon Dioxide in beta Carbonic Anhydrase | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase, ZINC ION | | Authors: | Aggarwal, M, Chua, T.K, Pinard, M.A, Szebenyi, D.M, McKenna, R. | | Deposit date: | 2015-05-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbon Dioxide "Trapped" in a beta-Carbonic Anhydrase.
Biochemistry, 54, 2015
|
|
3BYR
 
 | | Mode of Action of a Putative Zinc Transporter CzrB (Zn form) | | Descriptor: | CzrB protein, ZINC ION | | Authors: | Cherezov, V, Srinivasan, V, Szebenyi, D.M.E, Caffrey, M. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Mode of Action of a Putative Zinc Transporter CzrB in Thermus thermophilus
Structure, 16, 2008
|
|
3BYP
 
 | | Mode of Action of a Putative Zinc Transporter CzrB | | Descriptor: | CzrB protein, SULFATE ION | | Authors: | Cherezov, V, Srinivasan, V, Szebenyi, D.M.E, Caffrey, M. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the Mode of Action of a Putative Zinc Transporter CzrB in Thermus thermophilus
Structure, 16, 2008
|
|
1R12
 
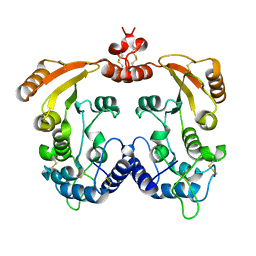 | | Native Aplysia ADP ribosyl cyclase | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R0S
 
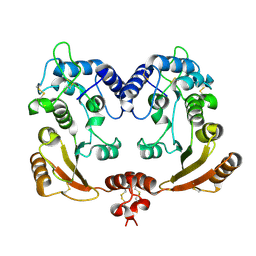 | | Crystal structure of ADP-ribosyl cyclase Glu179Ala mutant | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-22 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R16
 
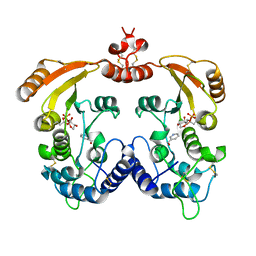 | | Aplysia ADP ribosyl cyclase with bound pyridylcarbinol and R5P | | Descriptor: | 3-PYRIDINYLCARBINOL, ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R15
 
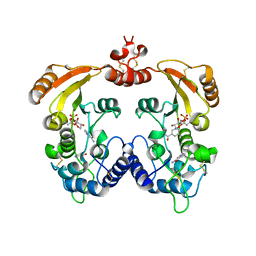 | | Aplysia ADP ribosyl cyclase with bound nicotinamide and R5P | | Descriptor: | ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE, NICOTINAMIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
3M7F
 
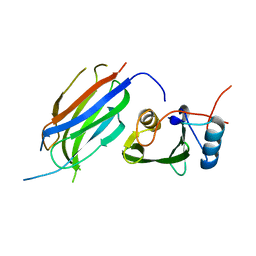 | | Crystal structure of the Nedd4 C2/Grb10 SH2 complex | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Growth factor receptor-bound protein 10 | | Authors: | Huang, Q, Szebenyi, M. | | Deposit date: | 2010-03-16 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Interaction between the Growth Factor-binding Protein GRB10 and the E3 Ubiquitin Ligase NEDD4.
J.Biol.Chem., 285, 2010
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
5T8B
 
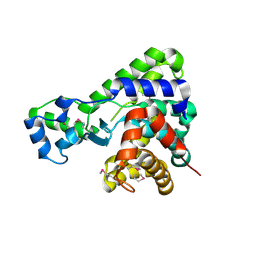 | |
5T8C
 
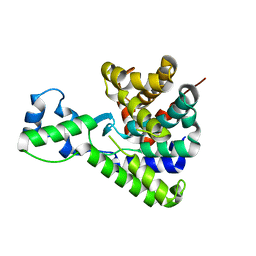 | |
1MHO
 
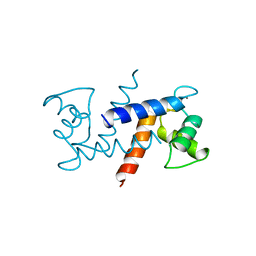 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
1B1G
 
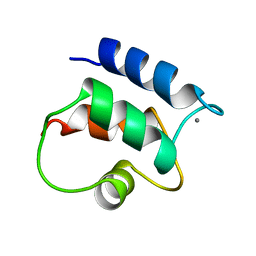 | |
6MUZ
 
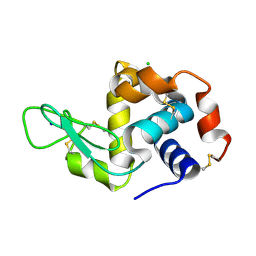 | | Lysozyme, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Kriksunov, I, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6MUY
 
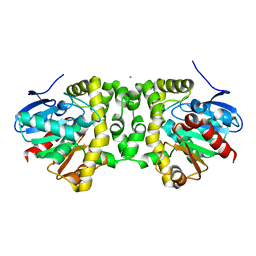 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6MV0
 
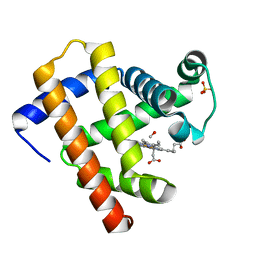 | | CO-bound Sperm Whale Myoglobin, room temperature structure solved by serial 5degree oscillation crystallography | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6MZZ
 
 | | Fluoroacetate dehalogenase, room temperature structure, using first 1 degree of total 3 degree oscillation | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6N03
 
 | | CO-bound Sperm Whale Myoglobin, room temperature structure, last 2 degrees of 5 degree total oscillation and 160 kGy dose | | Descriptor: | CARBON MONOXIDE, Myoglobin, CO-bound, ... | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
