5I87
 
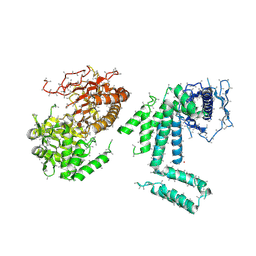 | | Crystal structure of BT-CD domains of human acetyl-CoA carboxylase | | Descriptor: | BT-CD domains of human acetyl-CoA carboxylase, CADMIUM ION | | Authors: | Stuttfeld, E, Hunkeler, M, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5T89
 
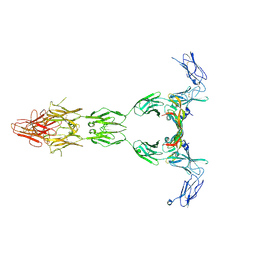 | | Crystal structure of VEGF-A in complex with VEGFR-1 domains D1-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vascular endothelial growth factor A, ... | | Authors: | Markovic-Mueller, S, Stuttfeld, E, Asthana, M, Weinert, T, Bliven, S, Goldie, K.N, Kisko, K, Capitani, G, Ballmer-Hofer, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A.
Structure, 25, 2017
|
|
4TM8
 
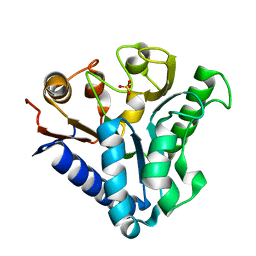 | |
4TM7
 
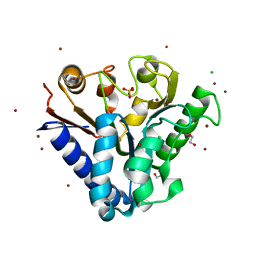 | | Crystal structure of 6-phosphogluconolactonase from Mycobacterium smegmatis N131D mutant soaked with CuSO4 | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphogluconolactonase, CHLORIDE ION, ... | | Authors: | Fujieda, N, Stuttfeld, E, Maier, T. | | Deposit date: | 2014-05-31 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Enzyme repurposing of a hydrolase as an emergent peroxidase upon metal binding.
Chem Sci, 6, 2015
|
|
4BSJ
 
 | | Crystal structure of VEGFR-3 extracellular domains D4-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 3 | | Authors: | Leppanen, V.-M, Tvorogov, D, Kisko, K, Prota, A.E, Jeltsch, M, Anisimov, A, Markovic-Mueller, S, Stuttfeld, E, Goldie, K.N, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mechanistic Insights Into Vegfr-3 Ligand Binding and Activation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BSK
 
 | | Crystal structure of VEGF-C in complex with VEGFR-3 domains D1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, VASCULAR ENDOTHELIAL GROWTH FACTOR C, ... | | Authors: | Leppanen, V.M, Tvorogov, D, Kisko, K, Prota, A.E, Jeltsch, M, Anisimov, A, Markovic-Mueller, S, Stuttfeld, E, Goldie, K.N, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural and Mechanistic Insights Into Vegfr-3 Ligand Binding and Activation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6G2H
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G2I
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) at 5.9 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1, Breast cancer type 1 susceptibility protein | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G2D
 
 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
5I6E
 
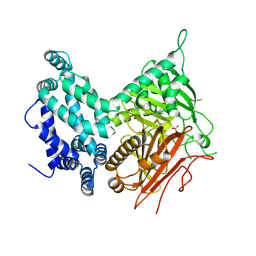 | | Crystal structure of the central domain of yeast acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase, MALONATE ION | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5FIF
 
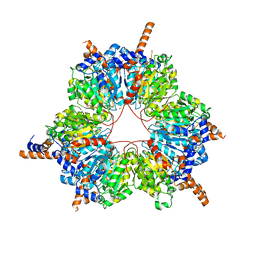 | | Carboxyltransferase domain of a single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
5I6F
 
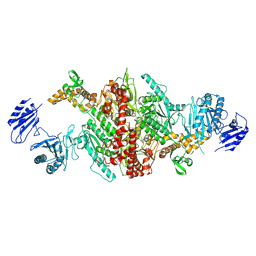 | | Crystal structure of C-terminal variant 1 of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5H80
 
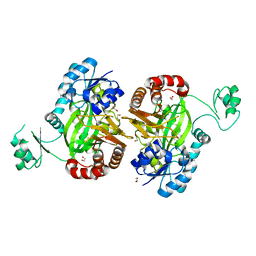 | | Biotin Carboxylase domain of single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
5I6G
 
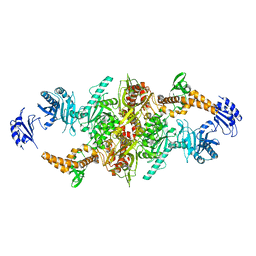 | | Crystal structure of C-terminal variant 2 of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5I6H
 
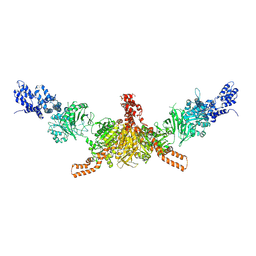 | | Crystal structure of CD-CT domains of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (7.2 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5I6I
 
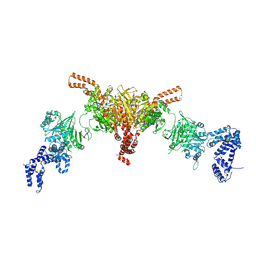 | | Crystal structure of a dBCCP-variant of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (8.4 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
6ZWO
 
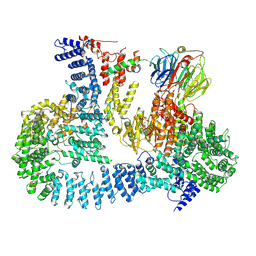 | | cryo-EM structure of human mTOR complex 2, focused on one half | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6ZWM
 
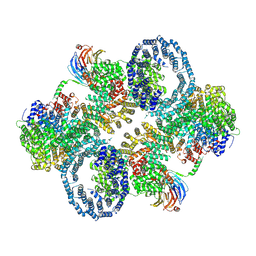 | | cryo-EM structure of human mTOR complex 2, overall refinement | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
7PEB
 
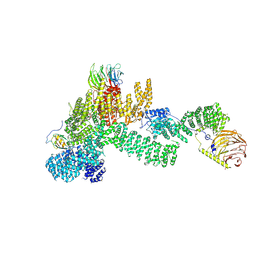 | |
7PEC
 
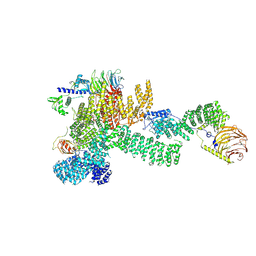 | |
7PED
 
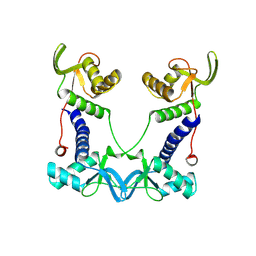 | | DEPTOR DEP domain tandem (DEPt) | | Descriptor: | DEP domain-containing mTOR-interacting protein | | Authors: | Waelchli, M, Jakob, R, Maier, T. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Regulation of human mTOR complexes by DEPTOR.
Elife, 10, 2021
|
|
7PE9
 
 | |
7PE7
 
 | |
7PE8
 
 | |
7PEA
 
 | |
