301D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | Descriptor: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
300D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
1MME
 
 | |
3ZD4
 
 | |
359D
 
 | |
4RJ1
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
4RKV
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
5UNE
 
 | |
2VRT
 
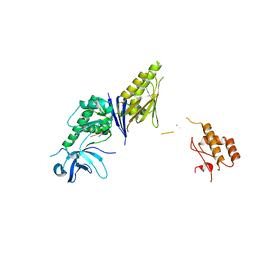 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
4RNE
 
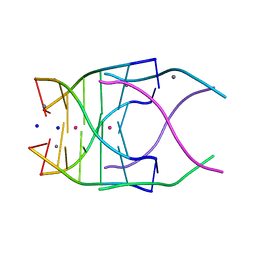 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
2OEU
 
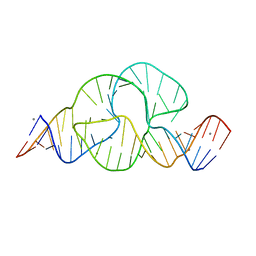 | | Full-length hammerhead ribozyme with Mn(II) bound | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*(OMC)P*CP*UP*GP*GP*(5BU)P*AP*UP*CP*CP*AP*AP*UP*CP*(DC))-3', Hammerhead Ribozyme, MANGANESE (II) ION | | Authors: | Martick, M, Scott, W.G. | | Deposit date: | 2007-01-01 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solvent structure and hammerhead ribozyme catalysis.
Chem.Biol., 15, 2008
|
|
1XJR
 
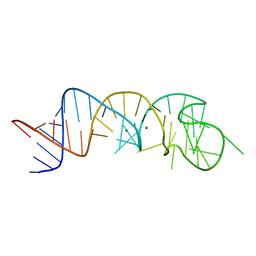 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | Descriptor: | MAGNESIUM ION, s2m RNA | | Authors: | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
379D
 
 | | THE STRUCTURAL BASIS OF HAMMERHEAD RIBOZYME SELF-CLEAVAGE | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*A P*GP*UP*CP*AP*CP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3') | | Authors: | Murray, J.B, Terwey, D.P, Maloney, L, Karpeisky, A, Usman, N, Beigelman, L, Scott, W.G. | | Deposit date: | 1998-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of hammerhead ribozyme self-cleavage.
Cell(Cambridge,Mass.), 92, 1998
|
|
488D
 
 | | CATALYTIC RNA ENZYME-PRODUCT COMPLEX | | Descriptor: | CADMIUM ION, FIRST RNA FRAGMENT OF CLEAVED SUBSTRATE, RNA RIBOZYME STRAND, ... | | Authors: | Murray, J.B, Szoke, H, Szoke, A, Scott, W.G. | | Deposit date: | 2000-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction.
Mol.Cell, 5, 2000
|
|
3ZP8
 
 | | HIGH-RESOLUTION FULL-LENGTH HAMMERHEAD RIBOZYME | | Descriptor: | HAMMERHEAD RIBOZYME, ENZYME STRAND, SUBSTRATE STRAND, ... | | Authors: | Anderson, M, Schultz, E, Martick, M, Scott, W.G. | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Monovalent Cations Revealed in a 1.55 A Resolution Hammerhead Ribozyme Structure
J.Mol.Biol., 425, 2013
|
|
3ZD5
 
 | |
2QUS
 
 | |
2QUW
 
 | |
2OIU
 
 | | L1 Ribozyme Ligase circular adduct | | Descriptor: | L1 Ribozyme RNA Ligase, MAGNESIUM ION | | Authors: | Robertson, M.P, Scott, W.G. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of ribozyme-catalyzed RNA assembly.
Science, 315, 2007
|
|
2VMK
 
 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
1Q29
 
 | |
1Y28
 
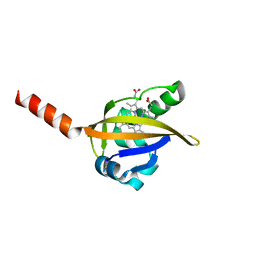 | | Crystal structure of the R220A metBJFIXL HEME domain | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Dunham, C.M, Dioum, E.M, Tuckerman, J.R, Gonzalez, G, Scott, W.G, Gilles-Gonzalez, M.A. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A distal arginine in the oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure
Biochemistry, 42, 2003
|
|
2C0B
 
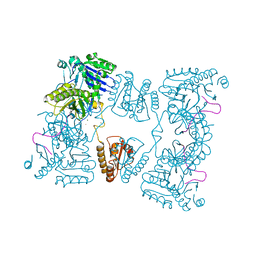 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2BX2
 
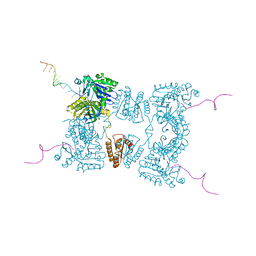 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, RNA (5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP* UP*GP*UP*U)-3'), ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
