1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
2RC6
 
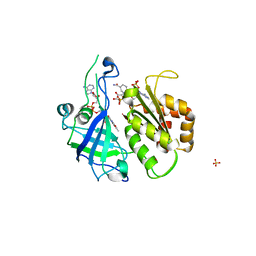 | | Refined structure of FNR from Leptospira interrogans bound to NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Ceccarelli, E.A, Polikarpov, I. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
5W0A
 
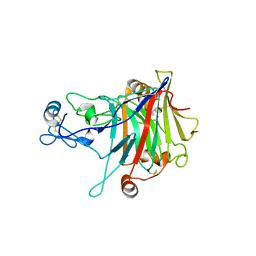 | | Crystal structure of Trichoderma harzianum endoglucanase I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucanase, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Godoy, A.S, Pellegrini, V.O.A, Sonoda, M.T, Kadowaki, M.A, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2017-05-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structure and dynamics of Trichoderma harzianum Cel7B suggest molecular architecture adaptations required for a wide spectrum of activities on plant cell wall polysaccharides.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5WAB
 
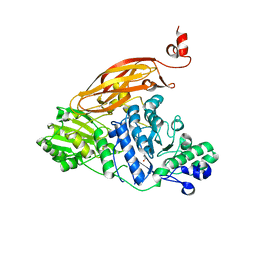 | |
4UZS
 
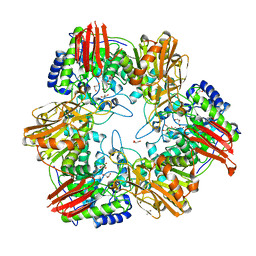 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpah-Galactose.
FEBS J., 283, 2016
|
|
4UOJ
 
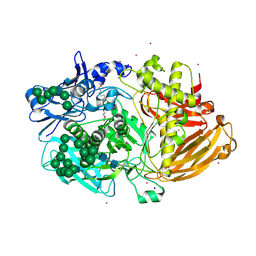 | | Structure of Fungal beta-mannosidase (GH2) from Trichoderma harzianum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Muniz, J.R.C, Aparicio, R, Santos, J.C, Nascimento, A.S, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Structure and Function of Fungal Beta-Mannosidases from Glycoside Hydrolase Family 2 Based on Multiple Crystal Structures of the Trichoderma Harzianum Enzyme.
FEBS J., 281, 2014
|
|
4USC
 
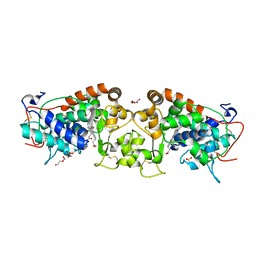 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
6O9N
 
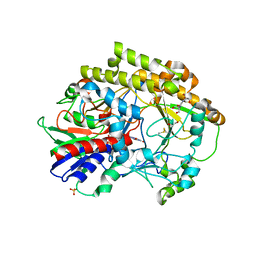 | | Structural insights on a new fungal aryl-alcohol oxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Enzymatic versatility and thermostability of a new aryl-alcohol oxidase from Thermothelomyces thermophilus M77.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
3CFN
 
 | |
3CFT
 
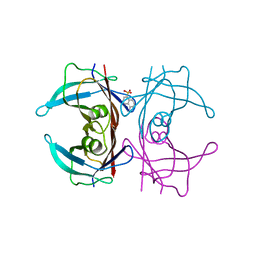 | |
3CFM
 
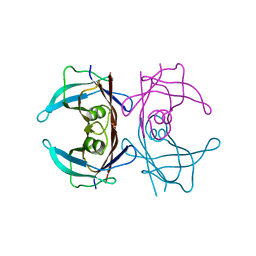 | |
5FLW
 
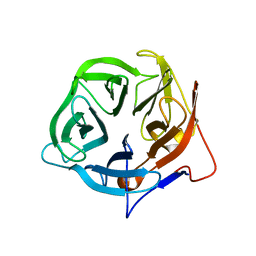 | | Crystal structure of putative exo-beta-1,3-galactanase from Bifidobacterium bifidum s17 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EXO-BETA-1,3-GALACTANASE | | Authors: | Godoy, A.S, de Lima, M.Z.T, Ramia, M.P, Camilo, C.M, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of a putative exo-beta-1,3-galactanase from Bifidobacterium bifidum S17.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4TUF
 
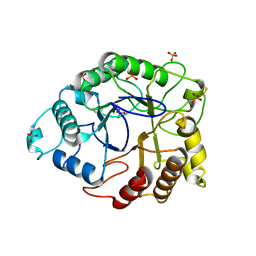 | |
5DYR
 
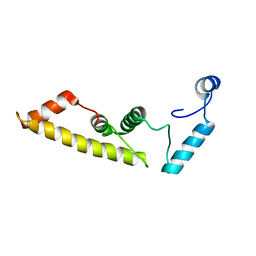 | | Structure of virulence-associated protein D (VapD) from Xylella fastidiosa | | Descriptor: | Virulence-associated protein D | | Authors: | Kochneva, M.V, dos Santos, M.L, dos Santos, C.A, de Souza, A.P, Polikarpov, I, Aparicio, R, Golubev, A.M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of virulence-associated protein D (VapD) from Xylella fastidiosa
To Be Published
|
|
6MVI
 
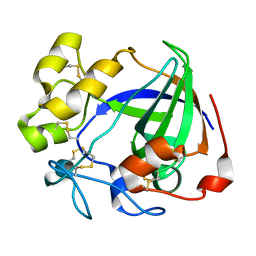 | | Apo Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
6MVJ
 
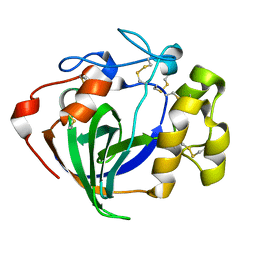 | | Cellobiose complex Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
4BCR
 
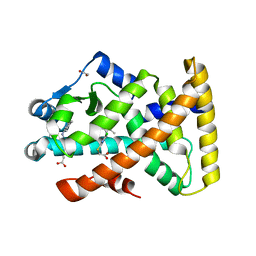 | | Structure of PPARalpha in complex with WY14643 | | Descriptor: | 1,2-ETHANEDIOL, 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR ALPHA | | Authors: | Bernardes, A, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2012-10-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Mechanism of Peroxisome Proliferator-Activated Receptor Alpha Activation by Wy14643: A New Mode of Ligand Recognition and Receptor Stabilization
J.Mol.Biol., 425, 2013
|
|
8GF3
 
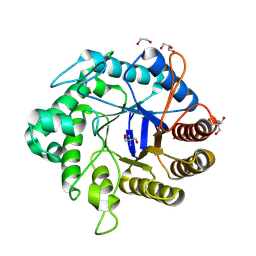 | | Crystallographic structure from BlMan5_7 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Briganti, L, Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-03-07 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic structure from BlMan5_7
To be published
|
|
2Y9N
 
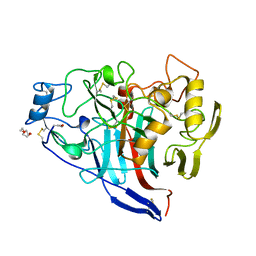 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EXOGLUCANASE 1, TRIETHYLENE GLYCOL | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2011-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cellobiohydrolase I from Trichoderma Harzianum: Structural and Enzymatic Characterization
To be Published
|
|
8U4F
 
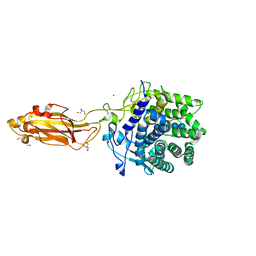 | |
8U4A
 
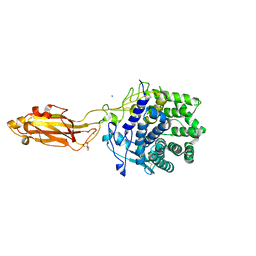 | |
8U49
 
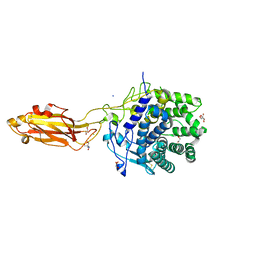 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8UWS
 
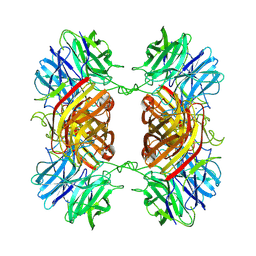 | | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43 | | Descriptor: | Beta-xylosidase, CALCIUM ION | | Authors: | Briganti, L, Godoy, A.S, Capetti, C.C.M, Portugal, R.V, Polikarpov, I. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43
To Be Published
|
|
6BUS
 
 | |
4UCF
 
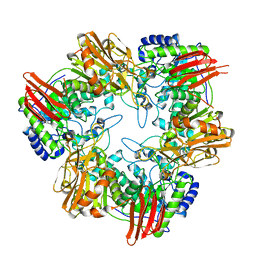 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
