2BNX
 
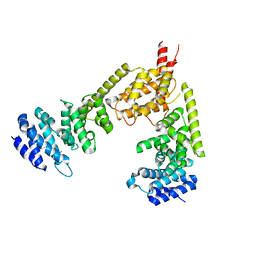 | | Crystal structure of the dimeric regulatory domain of mouse diaphaneous-related formin (DRF), mDia1 | | Descriptor: | CHLORIDE ION, DIAPHANOUS PROTEIN HOMOLOG 1 | | Authors: | Otomo, T, Otomo, C, Tomchick, D.R, Machius, M, Rosen, M.K. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Rho Gtpase-Mediated Activation of the Formin Mdia1
Mol.Cell, 18, 2005
|
|
1Y64
 
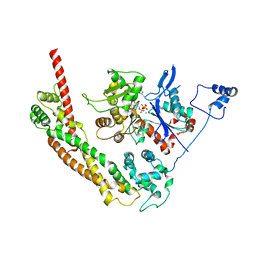 | | Bni1p Formin Homology 2 Domain complexed with ATP-actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Otomo, T, Tomchick, D.R, Otomo, C, Panchal, S.C, Machius, M, Rosen, M.K. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain
Nature, 433, 2005
|
|
1F2R
 
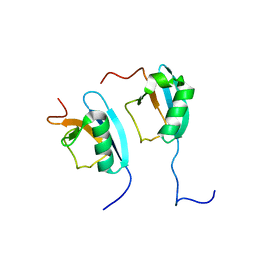 | | NMR STRUCTURE OF THE HETERODIMERIC COMPLEX BETWEEN CAD DOMAINS OF CAD AND ICAD | | Descriptor: | CASPASE-ACTIVATED DNASE, INHIBITOR OF CASPASE-ACTIVATED DNASE | | Authors: | Otomo, T, Sakahira, H, Uegaki, K, Nagata, S, Yamazaki, T. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the heterodimeric complex between CAD domains of CAD and ICAD.
Nat.Struct.Biol., 7, 2000
|
|
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
4NAW
 
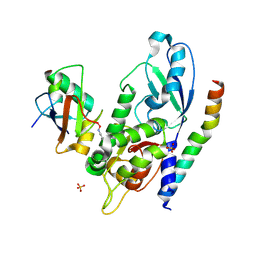 | | Crystal Structure of Human ATG12~ATG5-ATG16N in complex with a fragment of ATG3 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1, SODIUM ION, ... | | Authors: | Metlagel, Z, Otomo, C, Takaesu, G, Otomo, T. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GDL
 
 | | Crystal Structure of Human Atg12~Atg5 Conjugate in Complex with an N-terminal Fragment of Atg16L1 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1, SODIUM ION, ... | | Authors: | Otomo, C, Metlagel, Z, Takaesu, G, Otomo, T. | | Deposit date: | 2012-07-31 | | Release date: | 2012-12-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GDK
 
 | | Crystal Structure of Human Atg12~Atg5 Conjugate in Complex with an N-terminal Fragment of Atg16L1 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1, SODIUM ION, ... | | Authors: | Otomo, C, Metlagel, Z, Takaesu, G, Otomo, T. | | Deposit date: | 2012-07-31 | | Release date: | 2012-12-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7JLQ
 
 | | cryo-EM structure of human ATG9A in LMNG micelles | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLO
 
 | | Cryo-EM structure of human ATG9A in amphipols | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLP
 
 | | cryo-EM structure of human ATG9A in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7T5P
 
 | | Cryo-EM structure of human SIMC1-SLF2 complex | | Descriptor: | SMC5-SMC6 complex localization factor protein 2, SUMO-interacting motif-containing protein 1 | | Authors: | Maeda, S, Oravcova, M, Boddy, M.N, Otomo, T. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Nse5/6-like SIMC1-SLF2 complex localizes SMC5/6 to viral replication centers.
Elife, 11, 2022
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1C9F
 
 | |
7SWQ
 
 | |
7SVA
 
 | | Cryo-EM structure of Arabidopsis Ago10-guide RNA complex | | Descriptor: | MAGNESIUM ION, Protein argonaute 10, RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP*UP*UP*U)-3') | | Authors: | Xiao, Y, MacRae, I.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for RNA slicing by a plant Argonaute.
Nat.Struct.Mol.Biol., 30, 2023
|
|
