4RBQ
 
 | | 32 base pair oligo(U) RNA | | Descriptor: | POTASSIUM ION, U-Helix RNA from Trypanosome editing | | Authors: | Mooers, B.H.M. | | Deposit date: | 2014-09-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of the Trypanosome RNA Editing U-Helix with 16 Contiguous Us
To be Published
|
|
253D
 
 | |
339D
 
 | |
338D
 
 | |
340D
 
 | |
343D
 
 | |
345D
 
 | |
341D
 
 | |
342D
 
 | |
346D
 
 | |
337D
 
 | |
8G2K
 
 | |
7JU5
 
 | | Structure of RET protein tyrosine kinase in complex with pralsetinib | | Descriptor: | FORMIC ACID, Pralsetinib, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations.
Ann Oncol, 32, 2021
|
|
7JU6
 
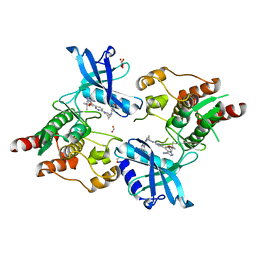 | | Structure of RET protein tyrosine kinase in complex with selpercatinib | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, Selpercatinib | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations.
Ann Oncol, 32, 2021
|
|
1FHZ
 
 | | PSORALEN CROSS-LINKED D(CCGGTACCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
1FHY
 
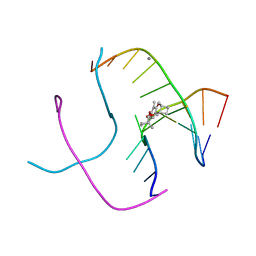 | | PSORALEN CROSS-LINKED D(CCGCTAGCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, CALCIUM ION, DNA (5'-D(*CP*CP*GP*CP*TP*AP*GP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
6NE7
 
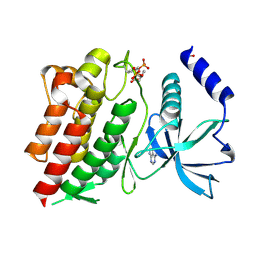 | | Structure of G810A mutant of RET protein tyrosine kinase domain. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NEC
 
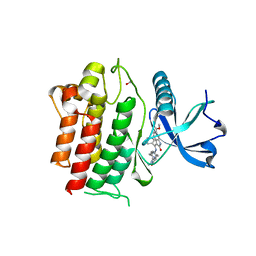 | | STRUCTURE OF RET PROTEIN TYROSINE KINASE DOMAIN IN COMPLEX WITH NINTEDANIB | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NJA
 
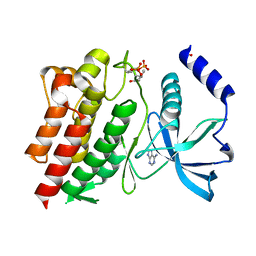 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
3C8R
 
 | |
3C7W
 
 | |
3C7Z
 
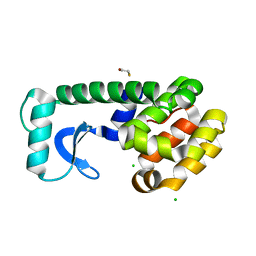 | | T4 lysozyme mutant D89A/R96H at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
5DA6
 
 | |
5D99
 
 | | 3DW4 redetermined by direct methods starting from random phase angles | | Descriptor: | GLYCEROL, RNA (27-MER) hairpin from sarcin-ricin domain of E. coli 23S rRNA | | Authors: | Mooers, B.H.M. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4PCO
 
 | |
