1B9Q
 
 | |
1B9P
 
 | |
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DNG
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL ACIDIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DN3
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
2K8J
 
 | | Solution structure of HCV p7 tm2 | | Descriptor: | p7tm2 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2008-09-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and ion channel activity of the p7 protein from hepatitis C virus.
J.Biol.Chem., 285, 2010
|
|
2N1P
 
 | |
2M5L
 
 | |
2LVG
 
 | |
2KWT
 
 | | Solution structure of NS2 [27-59] | | Descriptor: | Protease NS2-3 | | Authors: | Montserret, R, Bartenschlager, R, Penin, F. | | Deposit date: | 2010-04-19 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly.
Plos Pathog., 6, 2010
|
|
2KWZ
 
 | | Solution structure of NS2 [60-99] | | Descriptor: | Protease NS2-3 | | Authors: | Montserret, R, Bartenschlager, R, Penin, F. | | Deposit date: | 2010-04-22 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly.
Plos Pathog., 6, 2010
|
|
2JY0
 
 | |
2LZP
 
 | |
2JXF
 
 | | The solution structure of HCV NS4B(40-69) | | Descriptor: | Genome polyprotein | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2007-11-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Determinant for Membrane Association in Hepatitis C Virus Nonstructural Protein 4B
J.Virol., 83, 2009
|
|
2KZQ
 
 | | s34r Structure | | Descriptor: | Envelope glycoprotein E2 peptide | | Authors: | Montserret, R, Dubuisson, J, Penin, F. | | Deposit date: | 2010-06-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of new functional regions in hepatitis C virus envelope glycoprotein E2.
J.Virol., 85, 2011
|
|
2LIF
 
 | | Solution Structure of KKGF | | Descriptor: | Core protein p21 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2011-08-29 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of hepatitis C virus core-e1 signal Peptide and requirements for cleavage of the genotype 3a signal sequence by signal Peptide peptidase.
J.Virol., 86, 2012
|
|
2KDR
 
 | | Solution structure of HCV NS4B(227-254) | | Descriptor: | Non-structural protein 4B | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2009-01-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association
J.Virol., 83, 2009
|
|
2MKB
 
 | |
2LZQ
 
 | |
1CWX
 
 | | SOLUTION STRUCTURE OF THE HEPATITIS C VIRUS N-TERMINAL CAPSID PROTEIN 2-45 [C-HCV(2-45)] | | Descriptor: | HEPATITIS C VIRUS CAPSID PROTEIN | | Authors: | Ladaviere, L, Deleage, G, Montserret, R, Dalbon, P, Jolivet, M, Penin, F. | | Deposit date: | 1999-08-27 | | Release date: | 1999-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Immunodominant Antigenic Region of the Hepatitis C Virus Capsid Protein by NMR
To be Published
|
|
4WOE
 
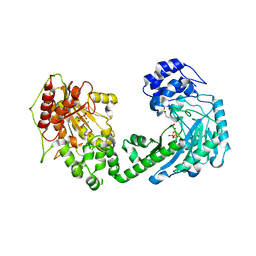 | | The duplicated taurocyamine kinase from Schistosoma mansoni with bound transition state analog (TSA) components | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
4WOD
 
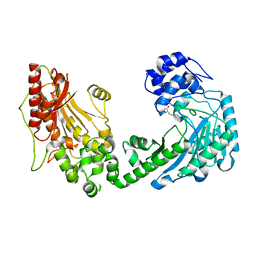 | | The duplicated taurocyamine kinase from Schistosoma mansoni complexed with arginine | | Descriptor: | ARGININE, Taurocyamine kinase | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
4WO8
 
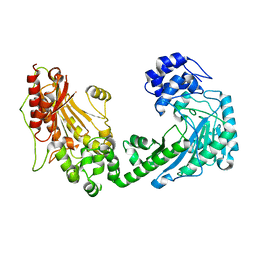 | | The substrate-free duplicated taurocyamine kinase from Schistosoma mansoni | | Descriptor: | Taurocyamine kinase | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
1UXC
 
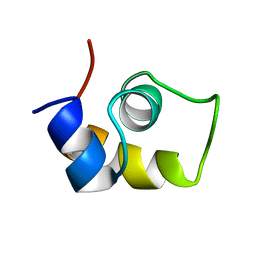 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1EMZ
 
 | | SOLUTION STRUCTURE OF FRAGMENT (350-370) OF THE TRANSMEMBRANE DOMAIN OF HEPATITIS C ENVELOPE GLYCOPROTEIN E1 | | Descriptor: | ENVELOPE GLYCOPROTEIN E1 | | Authors: | Op De Beeck, A, Montserret, R, Duvet, S, Cocquerel, L, Cacan, R, Barberot, B, Le Maire, M, Penin, F, Dubuisson, J. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization.
J.Biol.Chem., 275, 2000
|
|
