4RO1
 
 | |
5XMW
 
 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5C8X
 
 | | ZHD-Intermediate complex after ZHD crystal soaking in ZEN for 20min | | Descriptor: | (1E,10S)-1-(3,5-dihydroxyphenyl)-10-hydroxyundec-1-en-6-one, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Case Study of Lactonase ZHD
To Be Published
|
|
5C8Z
 
 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | Descriptor: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8JPP
 
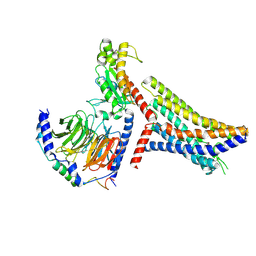 | | Cryo-EM structure of succinate receptor bound to succinate acid coupling MiniGsq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8K8J
 
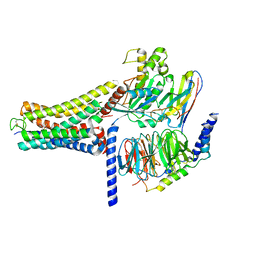 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
8JPN
 
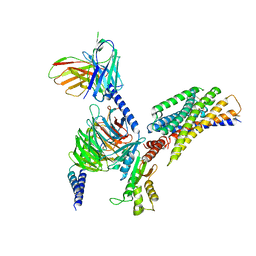 | | Cryo-EM structure of succinate receptor bound to cis-epoxysuccinic acid coupling to Gi | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6W41
 
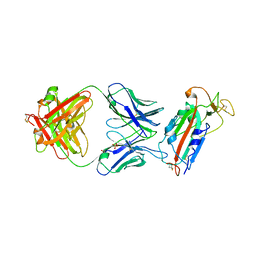 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Wu, N.C, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV.
Science, 368, 2020
|
|
5C7Y
 
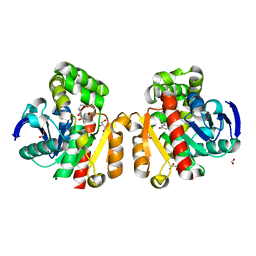 | | ZHD-Intermediate complex after ZHD crystal soaking in ZEN for 9min | | Descriptor: | (1E,10S)-1-(3,5-dihydroxyphenyl)-10-hydroxyundec-1-en-6-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Case Study of Lactonase ZHD
To Be Published
|
|
5C81
 
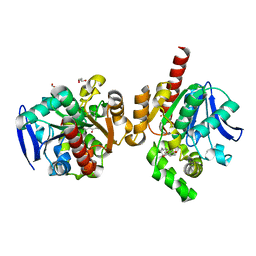 | | ZHD-Intermediate complex after ZHD crystal soaking in ZEN for 12min | | Descriptor: | (1E,10S)-1-(3,5-dihydroxyphenyl)-10-hydroxyundec-1-en-6-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Case Study of Lactonase ZHD
To Be Published
|
|
6NSB
 
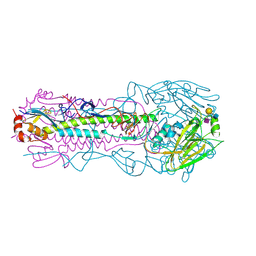 | |
6NSA
 
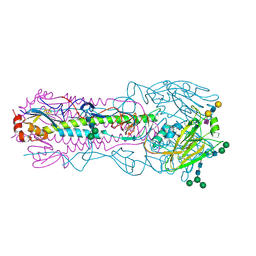 | |
6NSG
 
 | |
6NSC
 
 | | Crystal structure of the A/Brisbane/10/2007 (H3N2) influenza virus hemagglutinin G186V/L194P mutant apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6NS9
 
 | | Crystal structure of the IVR-165 (H3N2) influenza virus hemagglutinin apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6NSF
 
 | |
8EZ8
 
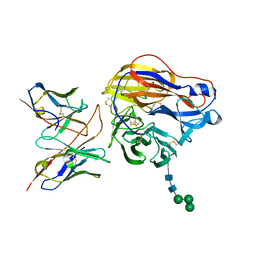 | | Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3C08, Light chain of influenza virus neuraminidase antibody 3C08, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ7
 
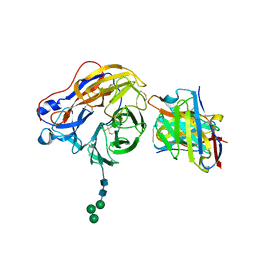 | | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 1F04, Light chain of influenza virus neuraminidase antibody 1F04, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ3
 
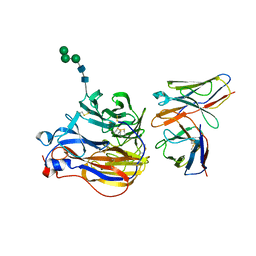 | | Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3A10, Light chain of influenza virus neuraminidase antibody 3A10, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
7CKB
 
 | | Simplified Alpha-Carboxysome, T=3 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
