7NY7
 
 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NY6
 
 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Histone macroH2A1.1 | | Authors: | Knobloch, G, Guberovic, I, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1EQF
 
 | | CRYSTAL STRUCTURE OF THE DOUBLE BROMODOMAIN MODULE FROM HUMAN TAFII250 | | Descriptor: | RNA POLYMERASE II TRANSCRIPTION INITIATION FACTOR, SULFATE ION | | Authors: | Jacobson, R.H, Ladurner, A.G, King, D.S, Tjian, R. | | Deposit date: | 2000-04-04 | | Release date: | 2000-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a human TAFII250 double bromodomain module.
Science, 288, 2000
|
|
3TE6
 
 | |
1ZR3
 
 | | Crystal structure of the macro-domain of human core histone variant macroH2A1.1 (form B) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, histone macroH2A1.1 | | Authors: | Kustatscher, G, Hothorn, M, Pugieux, C, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2005-05-19 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Splicing regulates NAD metabolite binding to histone macroH2A.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1ZR5
 
 | | Crystal structure of the macro-domain of human core histone variant macroH2A1.2 | | Descriptor: | H2AFY protein | | Authors: | Kustatscher, G, Hothorn, M, Pugieux, C, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Splicing regulates NAD metabolite binding to histone macroH2A.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BFR
 
 | | The Macro domain is an ADP-ribose binding module | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL PROTEIN AF1521, MAGNESIUM ION | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
2BFQ
 
 | | MACRO DOMAINS ARE ADP-RIBOSE BINDING MOLECULES | | Descriptor: | HYPOTHETICAL PROTEIN AF1521, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
3ZCO
 
 | | Crystal structure of S. cerevisiae Sir3 C-terminal domain | | Descriptor: | REGULATORY PROTEIN SIR3 | | Authors: | Oppikofer, M, Kueng, S, Keusch, J.J, Hassler, M, Ladurner, A.G, Gut, H, Gasser, S.M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of Sir3 Via its C-Terminal Winged Helix Domain is Essential for Yeast Heterochromatin Formation.
Embo J., 32, 2013
|
|
4AV1
 
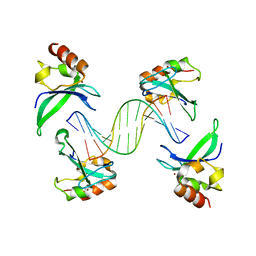 | | Crystal structure of the human PARP-1 DNA binding domain in complex with DNA | | Descriptor: | 5'-D(*AP*AP*GP*TP*GP*TP*TP*GP*CP*AP*TP*TP)-3', 5'-D(*TP*AP*AP*TP*GP*CP*AP*AP*CP*AP*CP*TP)-3', POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Ali, A.A.E, Timinszky, G, Arribas-Bosacoma, R, Kozlowski, M, Hassa, P.O, Hassler, M, Ladurner, A.G, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Zinc-Finger Domains of Parp1 Cooperate to Recognise DNA Strand-Breaks
Nat.Struct.Mol.Biol., 19, 2012
|
|
1H2G
 
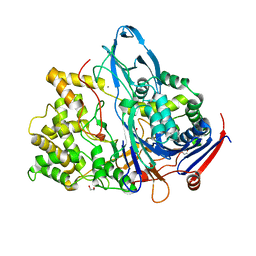 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
4KHA
 
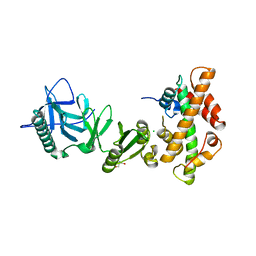 | | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Histone H2A, ... | | Authors: | Hondele, M, Halbach, F, Hassler, M, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
4KHO
 
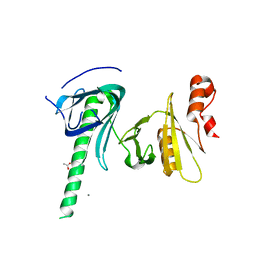 | |
4IQY
 
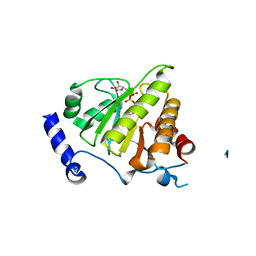 | | Crystal structure of the human protein-proximal ADP-ribosyl-hydrolase MacroD2 | | Descriptor: | MAGNESIUM ION, O-acetyl-ADP-ribose deacetylase MACROD2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Jankevicius, G, Hassler, M, Golia, B, Rybin, V, Zacharias, M, Timinszky, G, Ladurner, A.G. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A family of macrodomain proteins reverses cellular mono-ADP-ribosylation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KHB
 
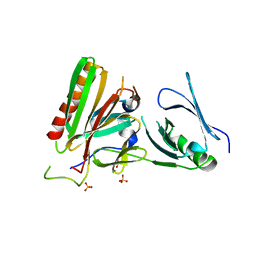 | | Structure of the Spt16D Pob3N heterodimer | | Descriptor: | SULFATE ION, Uncharacterized protein POB3N, Uncharacterized protein SPT16D | | Authors: | Stuwe, T, Zhang, E, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
2FXK
 
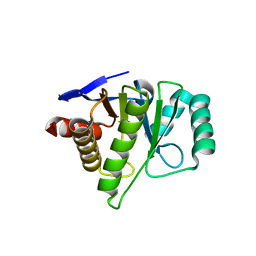 | | Crystal structure of the macro-domain of human core histone variant macroH2A1.1 (form A) | | Descriptor: | H2A histone family, member Y isoform 1 | | Authors: | Kustatscher, G, Hothorn, M, Pugieux, C, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2006-02-06 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Splicing regulates NAD metabolite binding to histone macroH2A.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3CB5
 
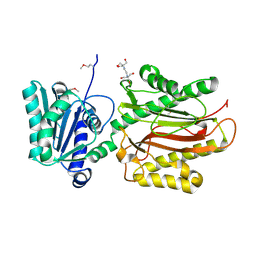 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form A) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CB6
 
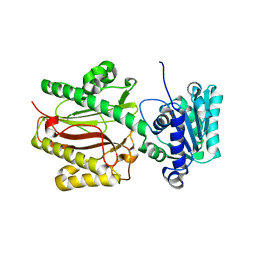 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3IIF
 
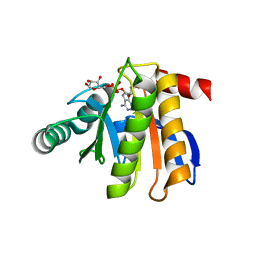 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form B) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1 | | Authors: | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3IID
 
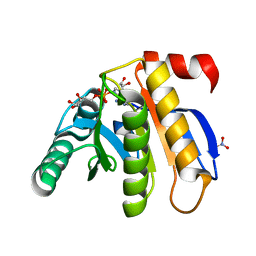 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form A) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1, ... | | Authors: | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3Q6Z
 
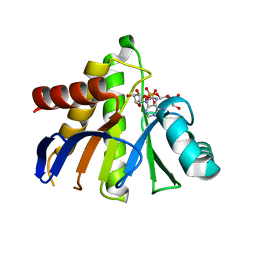 | | HUman PARP14 (ARTD8)-Macro domain 1 in complex with adenosine-5-diphosphoribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3Q71
 
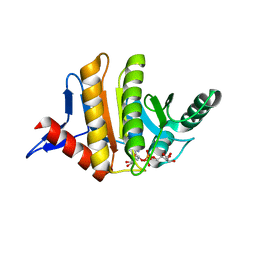 | | Human parp14 (artd8) - macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
1CIR
 
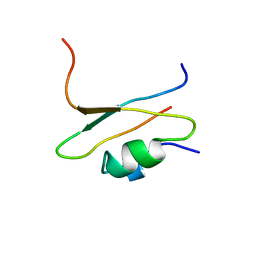 | | COMPLEX OF TWO FRAGMENTS OF CI2 [(1-40)(DOT)(41-64)] | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Davis, B.J, Fersht, A.R. | | Deposit date: | 1995-10-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards the complete structural characterization of a protein folding pathway: the structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism
Structure Fold.Des., 1, 1996
|
|
3V2B
 
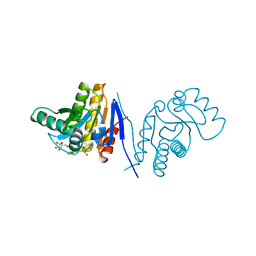 | | Human poly(adp-ribose) polymerase 15 (ARTD7, BAL3), macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 15, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Kotzcsh, A, Kraulis, P, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van den berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3VFQ
 
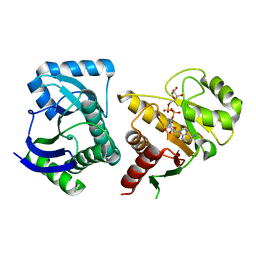 | | Human PARP14 (ARTD8, BAL2) - macro domains 1 and 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
