5Y0C
 
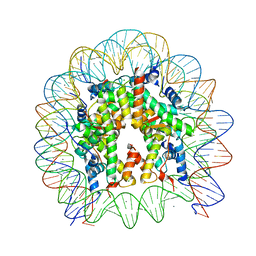 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y0D
 
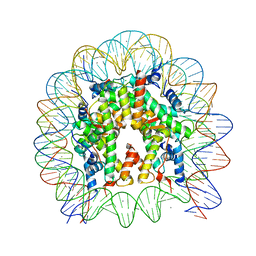 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
8YNY
 
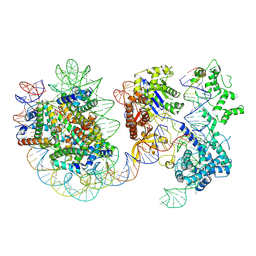 | | Structure of Cas9-sgRNA ribonucleoprotein bound to nucleosome | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (17-mer), DNA (175-mer), ... | | Authors: | Nagamura, R, Kujirai, T, Kusakizako, T, Hirano, H, Kurumizaka, H, Nureki, O. | | Deposit date: | 2024-03-12 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural insights into how Cas9 targets nucleosomes.
Nat Commun, 15, 2024
|
|
7YOZ
 
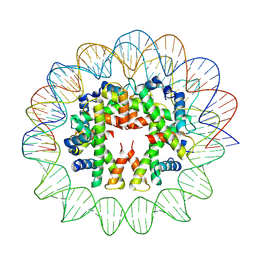 | | Cryo-EM structure of human subnucleosome (intermediate form) | | Descriptor: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YM6
 
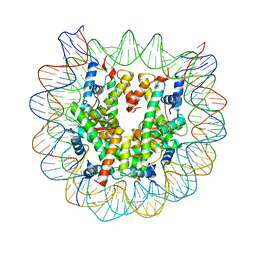 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4YM5
 
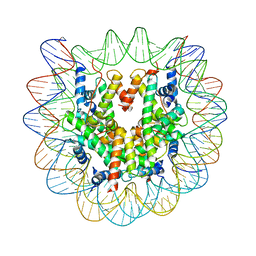 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
6V2K
 
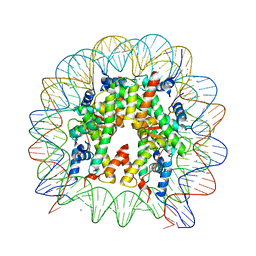 | | The nucleosome structure after H2A-H2B exchange | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Arimura, Y, Hirano, R, Kurumizaka, H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome.
Commun Biol, 4, 2021
|
|
5GXQ
 
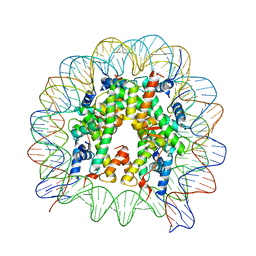 | | The crystal structure of the nucleosome containing H3.6 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
5GSE
 
 | | Crystal structure of unusual nucleosome | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kato, D, Osakabe, A, Arimura, Y, Park, S.Y, Kurumizaka, H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the overlapping dinucleosome composed of hexasome and octasome
Science, 356, 2017
|
|
5X7X
 
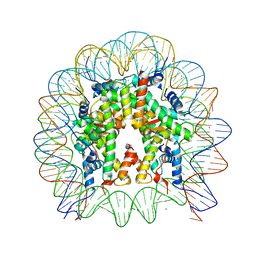 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
5XM1
 
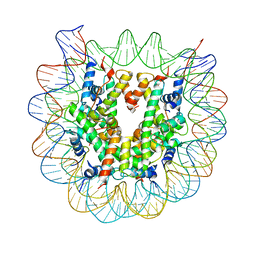 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5XM0
 
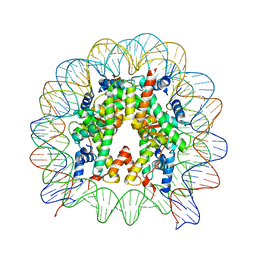 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
2EO0
 
 | |
5XRZ
 
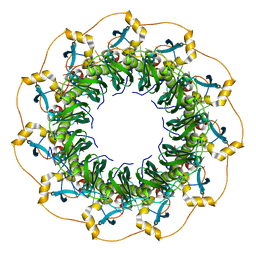 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
5XS0
 
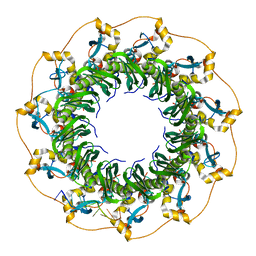 | | Structure of a ssDNA bound to the outer DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, ssDNA (5'-D(*CP*CP*CP*CP*CP*C)-3'), ssDNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
1AA3
 
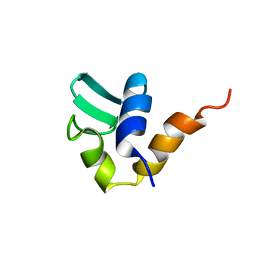 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RECA | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
1B22
 
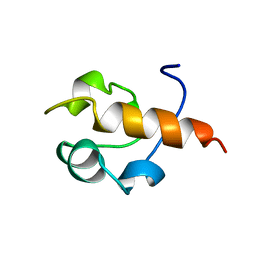 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
8H0W
 
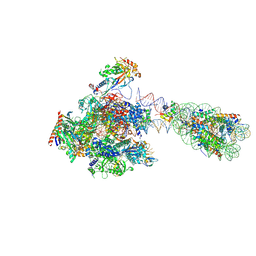 | | RNA polymerase II transcribing a chromatosome (type II) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8H0V
 
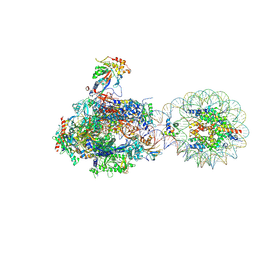 | | RNA polymerase II transcribing a chromatosome (type I) | | Descriptor: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2022-09-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8HE5
 
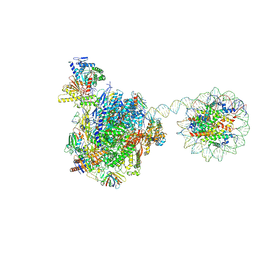 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8IHL
 
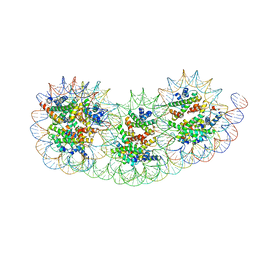 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
7XSE
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC42) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
