8IGU
 
 | |
8IGW
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
7COQ
 
 | |
6LLQ
 
 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BQS
 
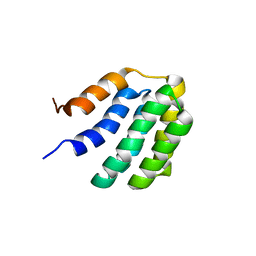 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Nomur | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Mussoc | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQQ
 
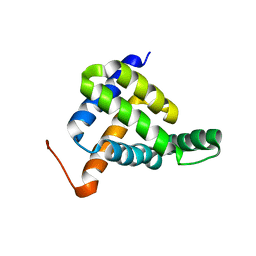 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Gogy | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
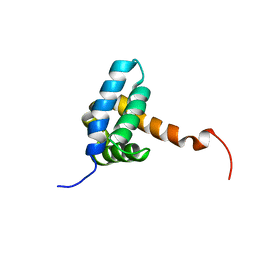 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Chantal | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Rei | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7DNS
 
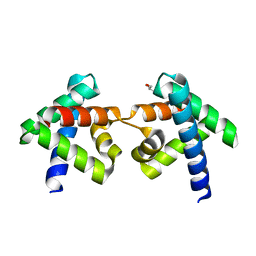 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
3WBL
 
 | | Crystal structure of CDK2 in complex with pyrazolopyrimidine inhibitor | | Descriptor: | ACETATE ION, Cyclin-dependent kinase 2, N~7~-(4-ethoxyphenyl)-6-methyl-N~5~-[(3S)-piperidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine | | Authors: | Fujino, A, Fukushima, K, Kubota, T, Kosugi, T, Takimoto-Kamimura, M. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase-2 complex with MK2 inhibitor TEI-I01800: insight into the selectivity.
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3A2C
 
 | | Crystal structure of a pyrazolopyrimidine inhibitor complex bound to MAPKAP Kinase-2 (MK2) | | Descriptor: | MAP kinase-activated protein kinase 2, N~7~-(4-ethoxyphenyl)-6-methyl-N~5~-[(3S)-piperidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine, SULFATE ION | | Authors: | Fujino, A, Takimoto-Kamimura, M. | | Deposit date: | 2009-05-12 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of an MK2-inhibitor complex: insight into the regulation of the secondary structure of the Gly-rich loop by TEI-I01800
Acta Crystallogr.,Sect.D, 66, 2010
|
|
