6E8H
 
 | | Legionella Longbeachae LeSH (Llo2327) | | Descriptor: | CHLORIDE ION, LeSH (Llo2327) | | Authors: | Kaneko, T, Li, S.S.C. | | Deposit date: | 2018-07-29 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and characterization of a large family of superbinding bacterial SH2 domains.
Nat Commun, 9, 2018
|
|
6E8K
 
 | |
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
6E8M
 
 | |
6E8I
 
 | |
4F59
 
 | | Triple mutant Src SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5A
 
 | | Triple mutant Src SH2 domain bound to phosphate ion | | Descriptor: | PHOSPHATE ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5B
 
 | | Triple mutant Src SH2 domain bound to phosphotyrosine | | Descriptor: | O-PHOSPHOTYROSINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
1UJ0
 
 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | Descriptor: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | Authors: | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | Deposit date: | 2003-07-24 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
1WPR
 
 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1WOM
 
 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
6DM4
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide | | Descriptor: | RavO, SULFATE ION, Shc1 phospho-Tyr317 peptide | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide
To Be Published
|
|
6DM3
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | Descriptor: | RavO | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
1VBU
 
 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1VBR
 
 | | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose | | Descriptor: | ACETIC ACID, alpha-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase B | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1RW9
 
 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
7SFR
 
 | | Unmethylated Mtb Ribosome 50S with SEQ-9 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Xing, Z, Cui, Z, Zhang, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-10-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7KGB
 
 | |
4KW5
 
 | | M. tuberculosis DprE1 in complex with inhibitor TCA1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Identification of a small molecule with activity against drug-resistant and persistent tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
1RWF
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWG
 
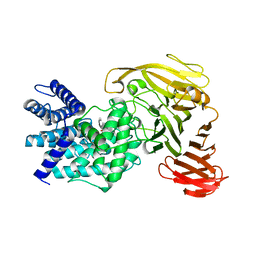 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWH
 
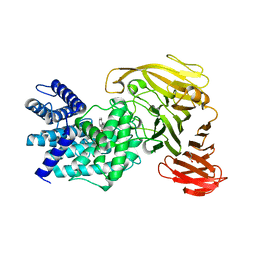 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWA
 
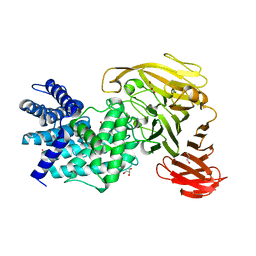 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | GLYCEROL, MERCURY (II) ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWC
 
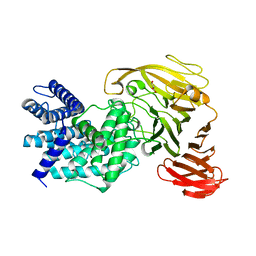 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
