1T73
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxFF motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxFF motif peptide, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T76
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a WxxVW motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7R
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLF motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLF motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T74
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a WxxLF motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7T
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with 5-alpha dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T79
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLW motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLW motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7F
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a LxxLL motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, LxxLL motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
1T7M
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxYF motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
4AE8
 
 | | Crystal structure of human THEM4 | | Descriptor: | THIOESTERASE SUPERFAMILY MEMBER 4 | | Authors: | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
4AE7
 
 | | Crystal structure of human THEM5 | | Descriptor: | THIOESTERASE SUPERFAMILY MEMBER 5 | | Authors: | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
1P0S
 
 | | Crystal Structure of Blood Coagulation Factor Xa in Complex with Ecotin M84R | | Descriptor: | Coagulation factor X precursor, Ecotin precursor, MAGNESIUM ION, ... | | Authors: | Wang, S.X, Hur, E, Sousa, C.A, Brinen, L, Slivka, E.J, Fletterick, R.J. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Extended Interactions and Gla Domain of Blood Coagulation Factor Xa
Biochemistry, 42, 2003
|
|
3J2I
 
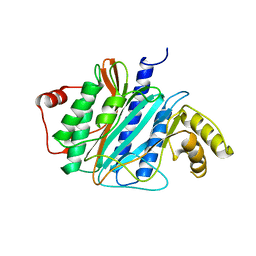 | | Structure of late pre-60S ribosomal subunits with nuclear export factor Arx1 bound at the peptide exit tunnel | | Descriptor: | Eukaryotic translation initiation factor 6, Proliferation-associated protein 2G4 | | Authors: | Bradatsch, B, Leidig, C, Granneman, S, Gnaedig, M, Tollervey, D, Boettcher, B, Beckmann, R, Hurt, E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
6EML
 
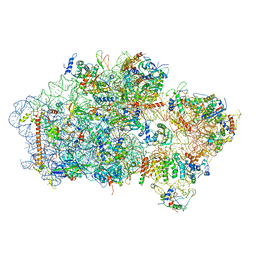 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
6RXU
 
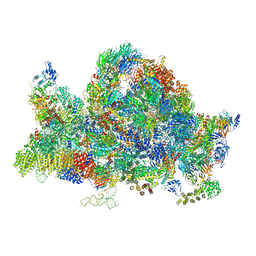 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6EM1
 
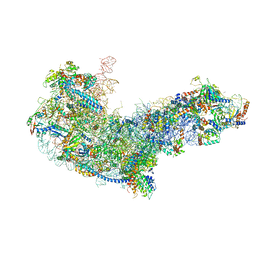 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6ELZ
 
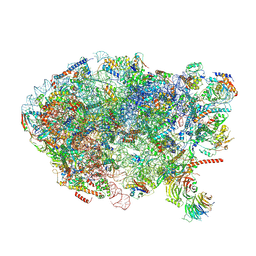 | | State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-09-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM3
 
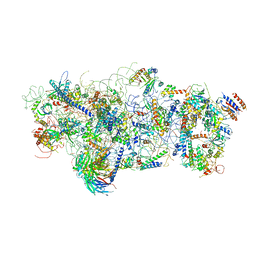 | | State A architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM4
 
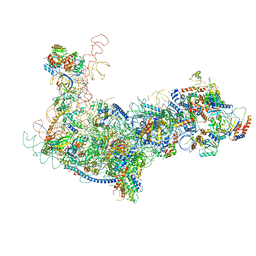 | | State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM5
 
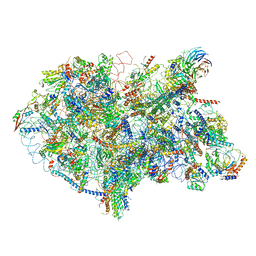 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
4WJS
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
3M99
 
 | | Structure of the Ubp8-Sgf11-Sgf73-Sus1 SAGA DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Kohler, A, Zimmerman, E, Schneider, M, Hurt, E, Zheng, N. | | Deposit date: | 2010-03-21 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module.
Cell(Cambridge,Mass.), 141, 2010
|
|
