1Z9C
 
 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
1Z91
 
 | |
3COQ
 
 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
4XRF
 
 | |
3UN9
 
 | | Crystal structure of an immune receptor | | Descriptor: | NLR family member X1, PLATINUM (II) ION | | Authors: | Hong, M, Yoon, S.I, Wilson, I.A. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Functional Characterization of the RNA-Binding Element of the NLRX1 Innate Immune Modulator.
Immunity, 36, 2012
|
|
4M5Z
 
 | | Crystal structure of broadly neutralizing antibody 5J8 bound to 2009 pandemic influenza hemagglutinin, HA1 subunit | | Descriptor: | Fab 5J8 heavy chain, Fab 5J8 light chain, Hemagglutinin HA1 chain | | Authors: | Hong, M, Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
2KAD
 
 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
7EYC
 
 | | Crystal structure of Tau and acetylated tau peptide antigen | | Descriptor: | ACETYLATED TAU PEPTIDE, antibody, Heavy chain, ... | | Authors: | Hong, M, Park, J. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Monoclonal antibody Y01 prevents tauopathy progression induced by lysine 280-acetylated tau in cell and mouse models.
J.Clin.Invest., 133, 2023
|
|
7BWL
 
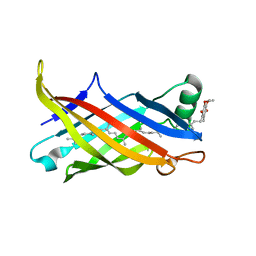 | | Structure of antibiotic sequester from Pseudomonas aerurinosa | | Descriptor: | UPF0312 protein PA0423, Ubiquinone-8 | | Authors: | Hong, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Pseudomonas aeruginosa PA0423 protein and its functional implication in antibiotic sequestration.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7WJP
 
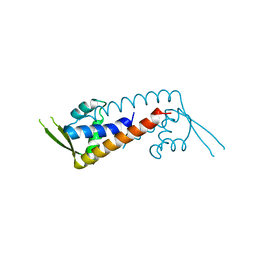 | | Structure of PadR-like protein from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Hong, M, Kim, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based molecular characterization of the LltR transcription factor from Listeria monocytogenes.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
7WZE
 
 | |
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
9CGZ
 
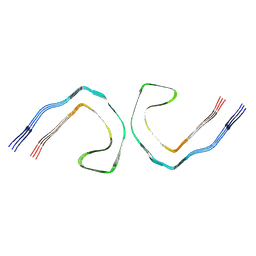 | | Alzheimer's Disease Seeded Mixed 0N4R and 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
9CGX
 
 | | Alzheimer's Disease Seeded 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
8SUZ
 
 | | Open State of the SARS-CoV-2 Envelope Protein Transmembrane Domain, Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Medeiros-Silva, J, Dregni, A.J, Somberg, N.H, Hong, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure of the open SARS-CoV-2 E viroporin.
Sci Adv, 9, 2023
|
|
8TTN
 
 | | PHF1-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S396E, S400E, T403E, S404E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TTL
 
 | | AT8-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S202E, T205E, S208E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7K3G
 
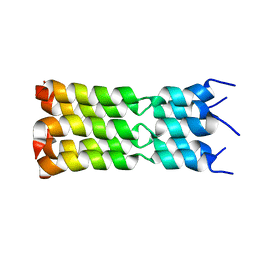 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Pentameric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Mandala, V.S, Hong, M, McKay, M.J, Shcherbakov, A.S, Dregni, A.J. | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JK8
 
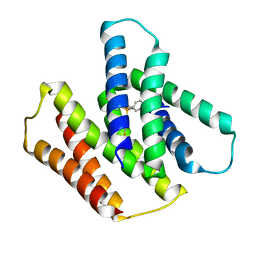 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
5ZQH
 
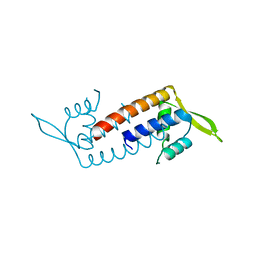 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
6PVR
 
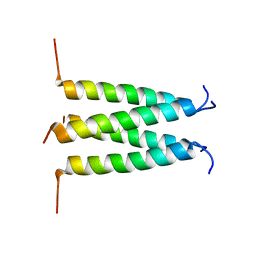 | | Influenza B M2 Proton Channel in the Closed State - SSNMR Structure at pH 7.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PVT
 
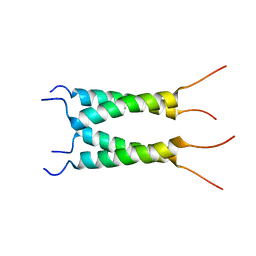 | | Influenza B M2 Proton Channel in the Open State - SSNMR Structure at pH 4.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8G54
 
 | |
8G55
 
 | |
8G58
 
 | |
