5B16
 
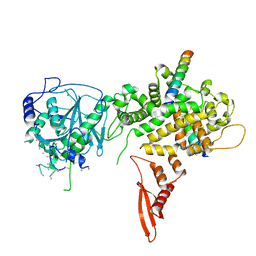 | | X-ray structure of DROSHA in complex with the C-terminal tail of DGCR8. | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3,DROSHA,Ribonuclease 3,DROSHA,Ribonuclease 3, ZINC ION | | Authors: | Kwon, S.C, Nguyen, T.A, Choi, Y.G, Jo, M.H, Hohng, S, Kim, V.N, Woo, J.S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human DROSHA
Cell, 164, 2016
|
|
6JM9
 
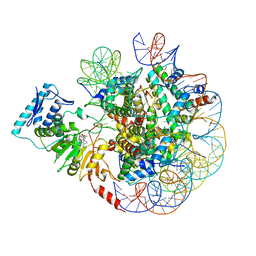 | | cryo-EM structure of DOT1L bound to unmodified nucleosome | | Descriptor: | DNA strand I, DNA strand J, Histone H2A, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
6JMA
 
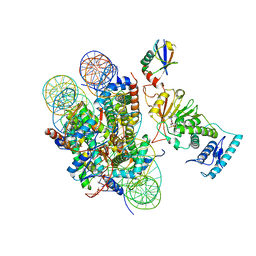 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
4ICS
 
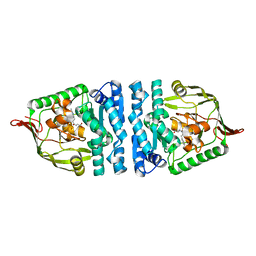 | | Crystal structure of PepS from Streptococcus pneumoniae in complex with a substrate | | Descriptor: | Aminopeptidase PepS, GLYCINE, TRYPTOPHAN, ... | | Authors: | Lee, S, Kim, K.K, Ta, M.H. | | Deposit date: | 2012-12-11 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ICR
 
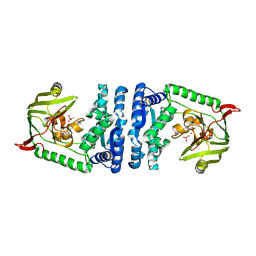 | | Structural basis for substrate recognition and reaction mechanism of bacterial aminopeptidase peps | | Descriptor: | Aminopeptidase PepS, CACODYLATE ION, ZINC ION | | Authors: | Lee, S, Kim, K.K, Ta, M.H. | | Deposit date: | 2012-12-11 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ICQ
 
 | |
4KMF
 
 | |
