1FOS
 
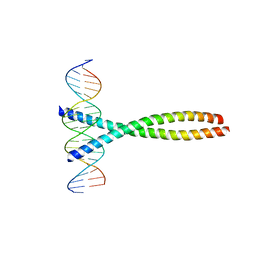 | | TWO HUMAN C-FOS:C-JUN:DNA COMPLEXES | | Descriptor: | C-JUN PROTO-ONCOGENE PROTEIN, DNA (5'-D(*AP*AP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*AP*T)-3'), ... | | Authors: | Glover, J.N.M, Harrison, S.C. | | Deposit date: | 1995-03-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA.
Nature, 373, 1995
|
|
3D8A
 
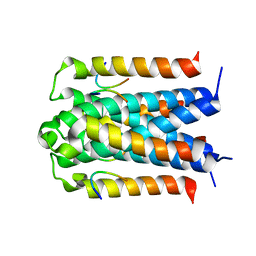 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
6DCX
 
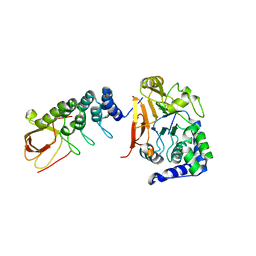 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
8UK7
 
 | |
5U6K
 
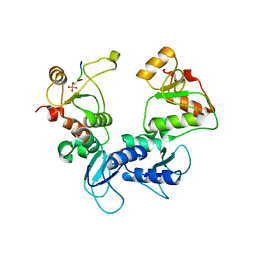 | |
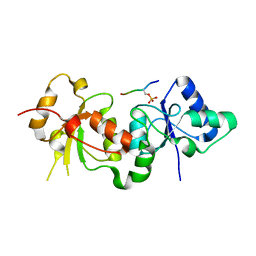
3PXE
 
 | |
2G9E
 
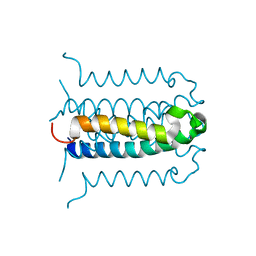 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TRAM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-03-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
3AL3
 
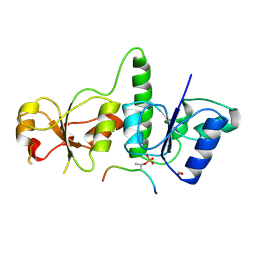 | | Crystal Structure of TopBP1 BRCT7/8-BACH1 peptide complex | | Descriptor: | DNA topoisomerase 2-binding protein 1, FORMIC ACID, Peptide of Fanconi anemia group J protein | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
3AL2
 
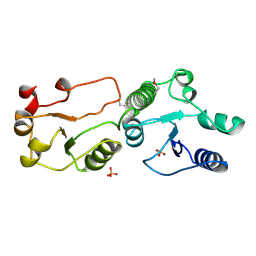 | | Crystal Structure of TopBP1 BRCT7/8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, SULFATE ION | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
3PXD
 
 | |
3PXA
 
 | |
3PXB
 
 | |
3PXC
 
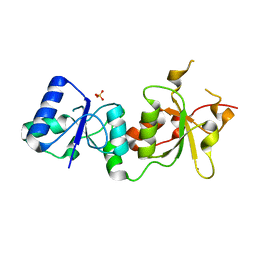 | |
3OMY
 
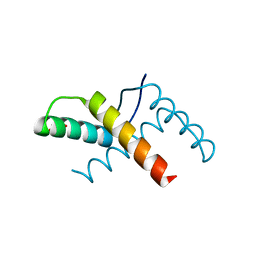 | | Crystal structure of the pED208 TraM N-terminal domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein traM | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
3ON0
 
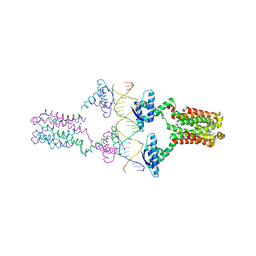 | | Crystal structure of the pED208 TraM-sbmA complex | | Descriptor: | Protein traM, sbmA | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
2RH3
 
 | | Crystal structure of plasmid pTiC58 VirC2 | | Descriptor: | Protein virC2 | | Authors: | Lu, J, Glover, J.N.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Agrobacterium tumefaciens VirC2 enhances T-DNA transfer and virulence through its C-terminal ribbon-helix-helix DNA-binding fold
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4WHV
 
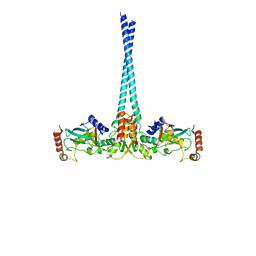 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
3UEO
 
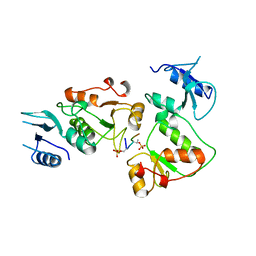 | |
3UEN
 
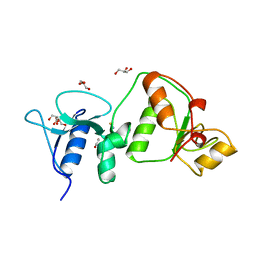 | | Crystal structure of TopBP1 BRCT4/5 domains | | Descriptor: | DNA topoisomerase 2-binding protein 1, GLYCEROL, THIOCYANATE ION | | Authors: | Leung, C.C, Glover, J.N.M. | | Deposit date: | 2011-10-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into recognition of MDC1 by TopBP1 in DNA replication checkpoint control.
Structure, 21, 2013
|
|
2ADO
 
 | | Crystal Structure Of The Brct Repeat Region From The Mediator of DNA damage checkpoint protein 1, MDC1 | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Lee, M.S, Edwards, R.A, Thede, G.L, Glover, J.N. | | Deposit date: | 2005-07-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the BRCT Repeat Domain of MDC1 and Its Specificity for the Free COOH-terminal End of the {gamma}-H2AX Histone Tail.
J.Biol.Chem., 280, 2005
|
|
4NRI
 
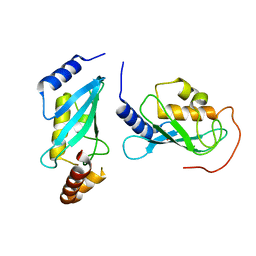 | |
4NR3
 
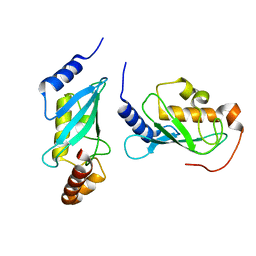 | |
4NRG
 
 | |
4ONM
 
 | | Crystal structure of human Mms2/Ubc13 - NSC697923 | | Descriptor: | 2-[(4-methylphenyl)sulfonyl]-5-nitrofuran, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
1YJ5
 
 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
