2WZO
 
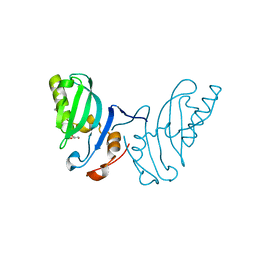 | | The structure of the FYR domain | | Descriptor: | GLYCEROL, TRANSFORMING GROWTH FACTOR BETA REGULATOR 1 | | Authors: | Garcia-Alai, M.M, Allen, M.D, Joerger, A.C, Bycroft, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Fyr Domain of Transforming Growth Factor Beta Regulator 1 (Tbrg1)(Pna)
Protein Sci., 19, 2010
|
|
7Q52
 
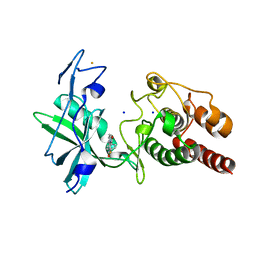 | | Crystal structure of S/T protein kinase PknG from Mycobacterium tuberculosis in complex with inhibitor L2W | | Descriptor: | 2-azanyl-3-(4-fluorophenyl)carbonyl-indolizine-1-carboxamide, FE (III) ION, SODIUM ION, ... | | Authors: | Defelipe, L.A, Burastero, O, Bento, I, Garcia-Alai, M.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cosolvent Sites-Based Discovery of Mycobacterium Tuberculosis Protein Kinase G Inhibitors.
J.Med.Chem., 65, 2022
|
|
6ENR
 
 | |
5ONF
 
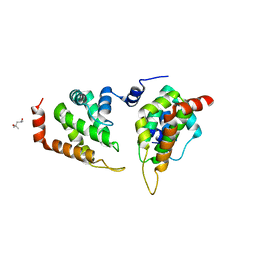 | | The ENTH domain from epsin-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epsin-1 | | Authors: | Garcia-Alai, M, GIeras, A, Meijers, R. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Epsin and Sla2 form assemblies through phospholipid interfaces.
Nat Commun, 9, 2018
|
|
5ON7
 
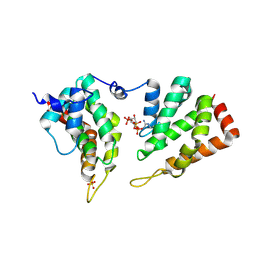 | | The ENTH domain from epsin-2 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | Epsin-2, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Garcia-Alai, M, GIeras, A, Meijers, R. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Epsin and Sla2 form assemblies through phospholipid interfaces.
Nat Commun, 9, 2018
|
|
5OO7
 
 | |
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7B2L
 
 | | Structure of the endocytic adaptor complex AENTH | | Descriptor: | ANTH domain of Sla2, ENTH domain of epsin Ent1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Klebl, D.P, Lizarrondo, J, Sobott, F, Garcia-Alai, M, Muench, S.P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the endocytic adaptor complex reveals the basis for efficient membrane anchoring during clathrin-mediated endocytosis.
Nat Commun, 12, 2021
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4MSL
 
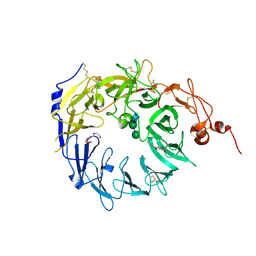 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF40431 | | Descriptor: | N-[(7-hydroxy-4-methyl-2-oxo-2H-chromen-8-yl)methyl]-L-leucine, Sortilin, TETRAETHYLENE GLYCOL, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the first small-molecule ligand of the neuronal receptor sortilin and structure determination of the receptor-ligand complex.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
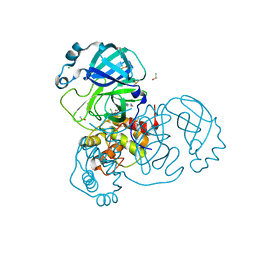 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ANS
 
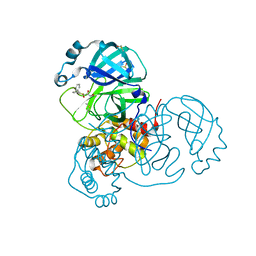 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | Descriptor: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AVD
 
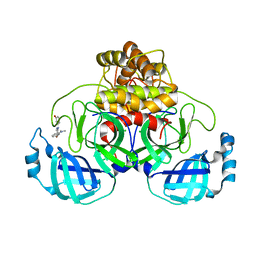 | | Structure of SARS-CoV-2 Main Protease bound to SEN1269 ligand | | Descriptor: | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol, 3C-like proteinase, CHLORIDE ION | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWU
 
 | | Structure of SARS-CoV-2 Main Protease bound to LSN2463359. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
