1HI9
 
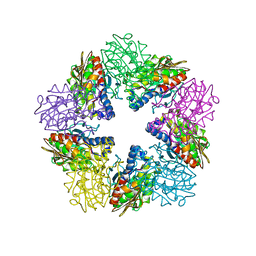 | | Zn-dependent D-aminopeptidase DppA from Bacillus subtilis, a self-compartmentalizing protease. | | Descriptor: | DIPEPTIDE TRANSPORT PROTEIN DPPA, ZINC ION | | Authors: | Remaut, H, Bompard-Gilles, C, Goffin, C, Frere, J.M, Van Beeumen, J. | | Deposit date: | 2001-01-04 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Bacillus Subtilis D-Aminopeptidase Dppa Reveals a Novel Self-Compartmentalizing Protease
Nat.Struct.Biol., 8, 2001
|
|
1B65
 
 | | Structure of l-aminopeptidase d-ala-esterase/amidase from ochrobactrum anthropi, a prototype for the serine aminopeptidases, reveals a new variant among the ntn hydrolase fold | | Descriptor: | PROTEIN (AMINOPEPTIDASE) | | Authors: | Bompard-Gilles, C, Villeret, V, Davies, G.J, Fanuel, L, Joris, B, Frere, J.M, Van Beeumen, J. | | Deposit date: | 1999-01-20 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new variant of the Ntn hydrolase fold revealed by the crystal structure
of L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi.
Structure Fold.Des., 8, 2000
|
|
1NRF
 
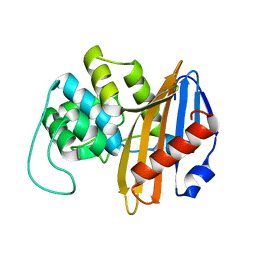 | | C-terminal domain of the Bacillus licheniformis BlaR penicillin-receptor | | Descriptor: | REGULATORY PROTEIN BLAR1 | | Authors: | Kerff, F, Charlier, P, Columbo, M.L, Sauvage, E, Brans, A, Frere, J.M, Joris, B, Fonze, E. | | Deposit date: | 2003-01-24 | | Release date: | 2004-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis.
Biochemistry, 42, 2003
|
|
1K38
 
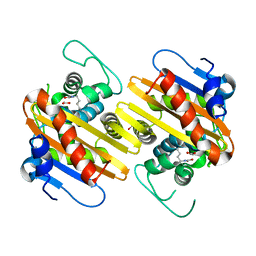 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1HIX
 
 | | CRYSTALLOGRAPHIC ANALYSES OF FAMILY 11 ENDO-BETA-1,4-XYLANASE XYL1 FROM STREPTOMYCES SP. S38 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Wouters, J, Georis, J, Dusart, J, Frere, J.M, Depiereux, E, Charlier, P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Analysis of Family 11 Endo-[Beta]-1,4-Xylanase Xyl1 from Streptomyces Sp. S38
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5W90
 
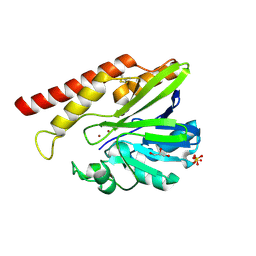 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5WCK
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
1W79
 
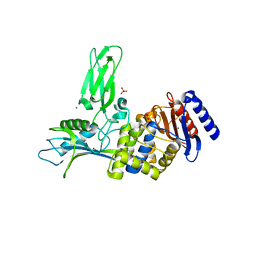 | | Crystal structure of the DD-transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins.
J. Biol. Chem., 280, 2005
|
|
1I2S
 
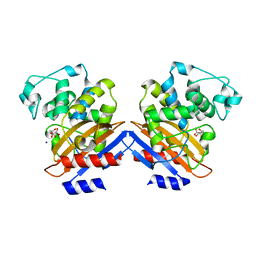 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 | | Descriptor: | BETA-LACTAMASE, CITRIC ACID, SODIUM ION | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the
acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1I2W
 
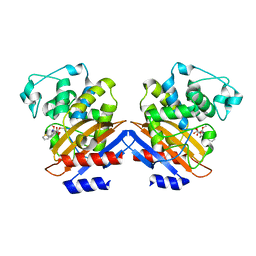 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1W5D
 
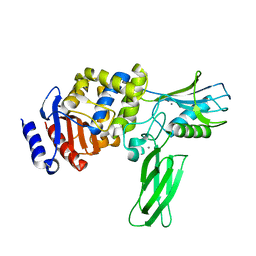 | | Crystal structure of PBP4a from Bacillus subtilis | | Descriptor: | CALCIUM ION, PENICILLIN-BINDING PROTEIN | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-06 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Penicillin-Binding Protein 4A, and its Complex with a Peptidoglycan Mimetic Peptide.
J.Mol.Biol., 371, 2007
|
|
3D2Z
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3D2Y
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
1BUE
 
 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
1K07
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | ACETATE ION, FEZ-1 beta-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2001-09-18 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-[beta]-lactamase from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.MOL.BIOL., 325, 2003
|
|
1K4F
 
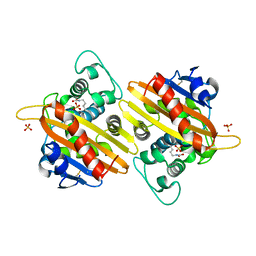 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.6 A RESOLUTION | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the class D beta-lactamase OXA-2
To be Published
|
|
1JT1
 
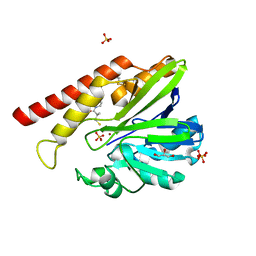 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, CHLORIDE ION, FEZ-1, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2001-08-20 | | Release date: | 2003-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-[beta]-lactamase from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.MOL.BIOL., 325, 2003
|
|
1K6R
 
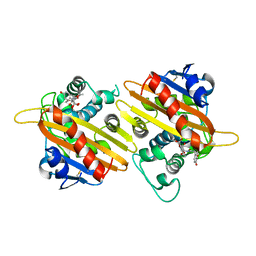 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH MOXALACTAM | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Beta-lactamase PSE-2 | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1K4E
 
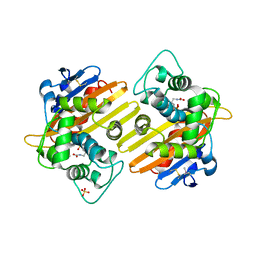 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K6S
 
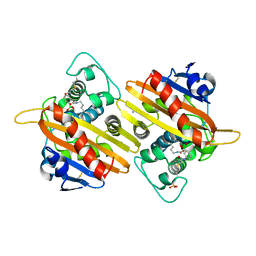 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH A PHENYLBORONIC ACID | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, Beta-lactamase PSE-2, CALCIUM ION, ... | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1DDK
 
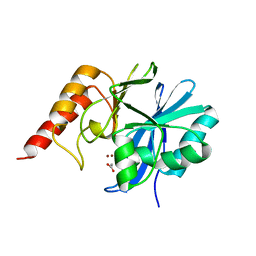 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DD6
 
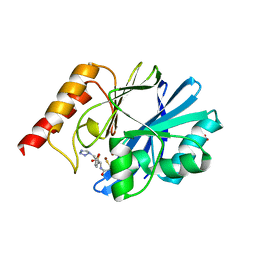 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1W8Y
 
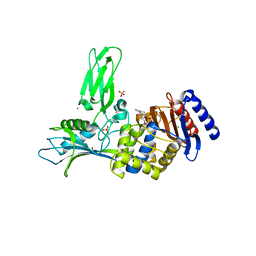 | | Crystal structure of the nitrocefin acyl-DD-peptidase from Actinomadura R39. | | Descriptor: | (2R)-2-{(1R)-2-OXO-1-[(2-THIENYLACETYL)AMINO]ETHYL}-5,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-10-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Actinomadura R39 Dd- Peptidase Reveals New Domains in Penicillin- Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
1W8Q
 
 | | Crystal Structure of the DD-Transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-09-24 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of the Actinomadura R39 Dd-Peptidase Reveals New Domains in Penicillin-Binding Proteins.
J.Biol.Chem., 280, 2005
|
|
3PTE
 
 | |
