1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1FB8
 
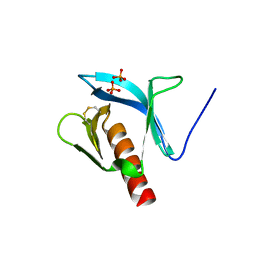 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, PHOSPHATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FHW
 
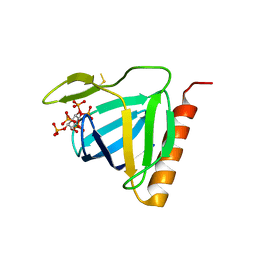 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1FAO
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-13 | | Release date: | 2000-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1MFG
 
 | |
1MFL
 
 | |
2MYW
 
 | |
2MYV
 
 | |
1EAZ
 
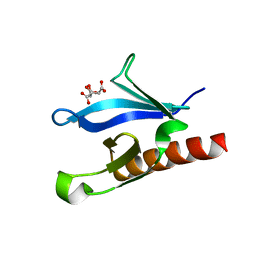 | | Crystal structure of the phosphoinositol (3,4)-bisphosphate binding PH domain of TAPP1 from human. | | Descriptor: | CITRIC ACID, TANDEM PH DOMAIN CONTAINING PROTEIN-1 | | Authors: | Thomas, C.C, Dowler, S, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Phosphatidylinositol 3,4-Bisphosphate-Binding Pleckstrin Homology (Ph) Domain of Tandem Ph-Domain-Containing Protein 1 (Tapp1): Molecular Basis of Lipid Specificity
Biochem.J., 358, 2001
|
|
