3NQ0
 
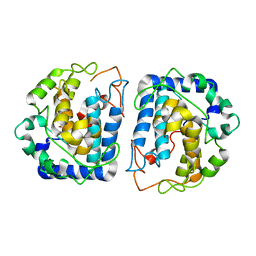 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of Zinc | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NM8
 
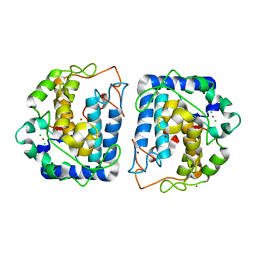 | | Crystal structure of Tyrosinase from Bacillus megaterium | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
3NTM
 
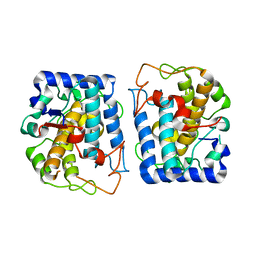 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of zinc, partial occupancy of CuB | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-07-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
3NPY
 
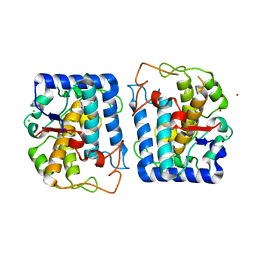 | | Crystal Structure of Tyrosinase from Bacillus megaterium soaked in CuSO4 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NQ1
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium in complex with inhibitor kojic acid | | Descriptor: | 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NQ5
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium R209H mutant | | Descriptor: | COPPER (II) ION, Tyrosinase, ZINC ION | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
6M5B
 
 | | X-ray crystal structure of cyclic-PIP and DNA complex in a reverse binding orientation | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[(4-azanyl-1-methyl-pyrrol-2-yl)carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-~{N}-[2-[[(3~{S})-3-azanyl-4-oxidanylidene-butyl]carbamoyl]-1-methyl-imidazol-4-yl]-1-methyl-imidazole-2-carboxamide, DNA (5'-D(*CP*(CBR)P*AP*GP*GP*CP*CP*TP*GP*G)-3'), ... | | Authors: | Abe, K, Hirose, Y, Eki, H, Takeda, K, Bando, T, Endo, M, Sugiyama, H. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | X-ray Crystal Structure of a Cyclic-PIP-DNA Complex in the Reverse-Binding Orientation.
J.Am.Chem.Soc., 142, 2020
|
|
1IPE
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1IPF
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH AND TROPINONE | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
3B40
 
 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
1OZI
 
 | | The alternatively spliced PDZ2 domain of PTP-BL | | Descriptor: | protein tyrosine phosphatase | | Authors: | Walma, T, Aelen, J, Oostendorp, M, van den Berk, L, Hendriks, W, Vuister, G.W. | | Deposit date: | 2003-04-09 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Closed Binding Pocket and Global Destabilization Modify the Binding Properties of an Alternatively Spliced Form of the Second PDZ Domain of PTP-BL.
Structure, 12, 2004
|
|
1ITV
 
 | | Dimeric form of the haemopexin domain of MMP9 | | Descriptor: | MMP9, SULFATE ION | | Authors: | Cha, H, Kopetzki, E, Huber, R, Lanzendoerfer, M, Brandstetter, H. | | Deposit date: | 2002-02-11 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the adaptive molecular recognition by MMP9.
J.Mol.Biol., 320, 2002
|
|
3BIL
 
 | | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum | | Descriptor: | Probable LacI-family transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum.
To be Published
|
|
2QU7
 
 | | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
2QSJ
 
 | | Crystal structure of a LuxR family DNA-binding response regulator from Silicibacter pomeroyi | | Descriptor: | DNA-binding response regulator, LuxR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a LuxR family DNA-binding response regulator from Silicibacter pomeroyi.
To be Published
|
|
2QR3
 
 | | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis | | Descriptor: | Two-component system response regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis.
To be Published
|
|
3T2N
 
 | | Human hepsin protease in complex with the Fab fragment of an inhibitory antibody | | Descriptor: | Antibody, Fab fragment, Heavy Chain, ... | | Authors: | Koschubs, T, Dengl, S, Duerr, H, Kaluza, K, Georges, G, Hartl, C, Jennewein, S, Lanzendoerfer, M, Auer, J, Stern, A, Huang, K.-S, Kostrewa, D, Ries, S, Hansen, S, Kohnert, U, Cramer, P, Mundigl, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric antibody inhibition of human hepsin protease.
Biochem.J., 442, 2012
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
3CTA
 
 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
7YI7
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-[4-(trifluoromethyl)phenyl]ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
7YJC
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
6KU3
 
 | | Crystal structure of gibberellin 2-oxidase3 (GA2ox3)in rice | | Descriptor: | 2-OXOGLUTARIC ACID, GIBBERELLIN A4, GLYCEROL, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
6KUN
 
 | | Crystal structure of dioxygenase for auxin oxidation (DAO) in rice | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent dioxygenase DAO, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
8JYG
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
