1U7M
 
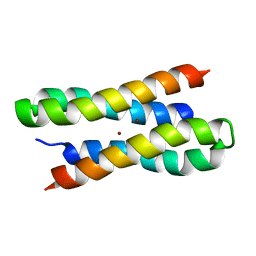 | | Solution structure of a diiron protein model: Due Ferri(II) turn mutant | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1U7J
 
 | | Solution structure of a diiron protein model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | Authors: | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-26 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
2HZ8
 
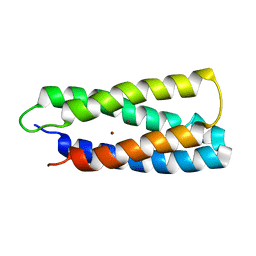 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
1NVO
 
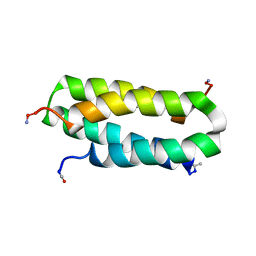 | | Solution structure of a four-helix bundle model, apo-DF1 | | Descriptor: | Homodimeric Alpha2 Four-Helix Bundle | | Authors: | Maglio, O, Nastri, F, Pavone, V, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2003-02-04 | | Release date: | 2003-03-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Preorganization of molecular binding sites in designed diiron proteins
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2A3D
 
 | | SOLUTION STRUCTURE OF A DE NOVO DESIGNED SINGLE CHAIN THREE-HELIX BUNDLE (A3D) | | Descriptor: | PROTEIN (DE NOVO THREE-HELIX BUNDLE) | | Authors: | Walsh, S.T.R, Cheng, H, Bryson, J.W, Roder, H, Degrado, W.F. | | Deposit date: | 1999-04-01 | | Release date: | 1999-05-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a de novo designed three-helix bundle protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7BEY
 
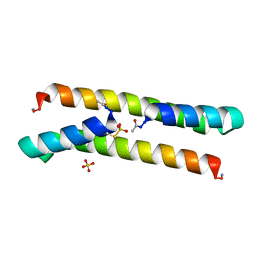 | | Het-N2-SO3- - De novo designed three-helix heterodimer with Cysteine S-sulfate at the N2 position of the alpha-helix | | Descriptor: | 'Cys-N2-SO3-' Strand, 'Positive' Strand, SULFATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
5WLK
 
 | |
6OUG
 
 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
6MCT
 
 | |
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | Descriptor: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | Descriptor: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
7JH6
 
 | | De novo designed two-domain di-Zn(II) and porphyrin-binding protein | | Descriptor: | NONAETHYLENE GLYCOL, Two-domain di-Zn(II) and porphyrin-binding protein, ZINC ION, ... | | Authors: | Schmidt, N, Liu, L, DeGrado, W.F. | | Deposit date: | 2020-07-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric cooperation in a de novo-designed two-domain protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TNB
 
 | |
8TNC
 
 | |
8TN1
 
 | |
8TND
 
 | |
8TN6
 
 | |
7UDV
 
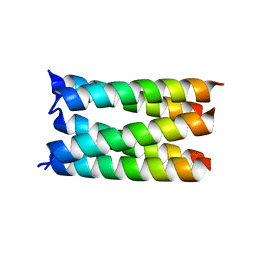 | |
7UDZ
 
 | | Designed pentameric proton channel LQLL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDW
 
 | | Designed pentameric proton channel QQLL | | Descriptor: | De novo designed pentameric proton channel QQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDY
 
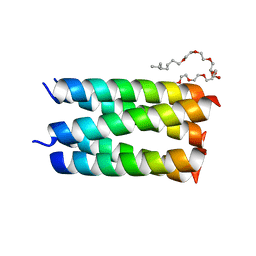 | | Designed pentameric channel QLLL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDX
 
 | | Designed pentameric proton channel QLQL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel QLQL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
3LBW
 
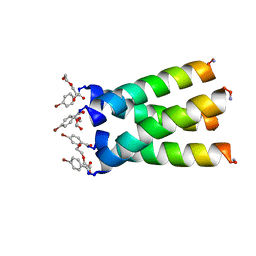 | | High resolution crystal structure of transmembrane domain of M2 | | Descriptor: | 4-bromobenzoic acid, DI(HYDROXYETHYL)ETHER, M2 protein, ... | | Authors: | Acharya, R, Polishchuk, A.L, DeGrado, W.F. | | Deposit date: | 2010-01-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
