2B4F
 
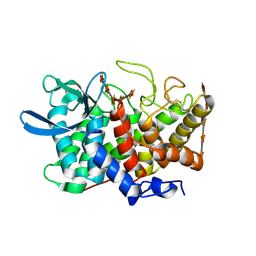 | | Structure Of A Cold-Adapted Family 8 Xylanase in complex with substrate | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Savvides, S.N, Feller, G, Van Beeumen, J.J. | | Deposit date: | 2005-09-23 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the beta-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product.
Biochemistry, 45, 2006
|
|
2BE7
 
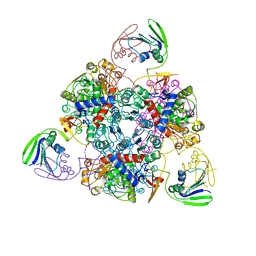 | | Crystal structure of the unliganded (T-state) aspartate transcarbamoylase of the psychrophilic bacterium Moritella profunda | | Descriptor: | Aspartate Carbamoyltransferase Catalytic Chain, Aspartate Carbamoyltransferase Regulatory Chain, SULFATE ION, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda.
J.Mol.Biol., 365, 2007
|
|
2BE9
 
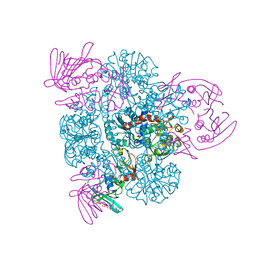 | | Crystal structure of the CTP-liganded (T-State) aspartate transcarbamoylase from the extremely thermophilic archaeon Sulfolobus acidocaldarius | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J.J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
Biochem.Biophys.Res.Commun., 372, 2008
|
|
1PG5
 
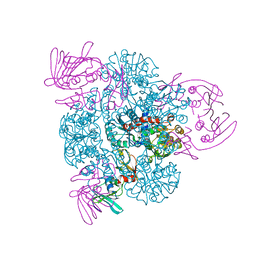 | | CRYSTAL STRUCTURE OF THE UNLIGATED (T-STATE) ASPARTATE TRANSCARBAMOYLASE FROM THE EXTREMELY THERMOPHILIC ARCHAEON SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | De Vos, D, Van Petegem, F, Remaut, H, Legrain, C, Glansdorff, N, Van Beeumen, J.J. | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of T State Aspartate Carbamoyltransferase of the Hyperthermophilic Archaeon Sulfolobus acidocaldarius.
J.Mol.Biol., 339, 2004
|
|
1XWQ
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
1XWT
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
2R9Z
 
 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
1XW2
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | Endo-1,4-beta-Xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2004-10-29 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
2A8Z
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2005-07-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase.
J.Mol.Biol., 354, 2005
|
|
3LN7
 
 | |
3LN6
 
 | |
6HXH
 
 | | Structure of the human ATP citrate lyase holoenzyme in complex with citrate, coenzyme A and Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase,Human ATP citrate lyase, CITRATE ANION, ... | | Authors: | Verstraete, K, Verschueren, K. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature, 568, 2019
|
|
6QCL
 
 | |
6QFB
 
 | |
6HXI
 
 | |
6HXL
 
 | |
6HXP
 
 | |
6HXJ
 
 | |
6HXN
 
 | |
6HXO
 
 | |
6HXQ
 
 | |
6HXK
 
 | |
6HXM
 
 | |
