7WOK
 
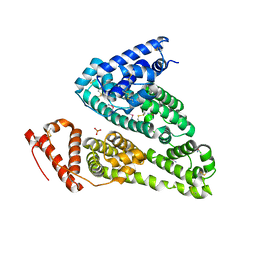 | | Crystal structure of HSA soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, PHOSPHATE ION | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
7WOJ
 
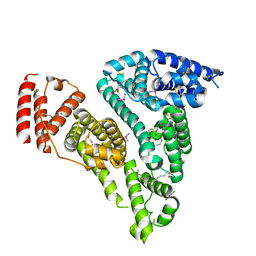 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
4PFE
 
 | | Crystal structure of vsfGFP-0 | | Descriptor: | Green fluorescent protein | | Authors: | Jauch, R, Chen, S.L. | | Deposit date: | 2014-04-29 | | Release date: | 2015-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Rational Structure-Based Design of Bright GFP-Based Complexes with Tunable Dimerization.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7TN9
 
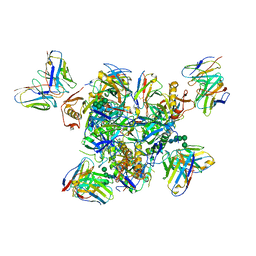 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
1D40
 
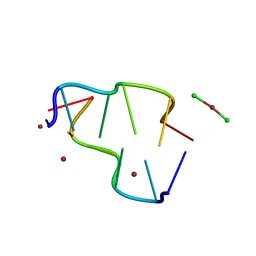 | | BASE SPECIFIC BINDING OF COPPER(II) TO Z-DNA: THE 1.3-ANGSTROMS SINGLE CRYSTAL STRUCTURE OF D(M5CGUAM5CG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) CHLORIDE, COPPER (II) ION, DNA (5'-D(*(5CM)P*(CU)GP*UP*AP*(5CM)P*(CU)G)-3') | | Authors: | Geierstanger, B.H, Kagawa, T.F, Chen, S.-L, Quigley, G.J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
1D32
 
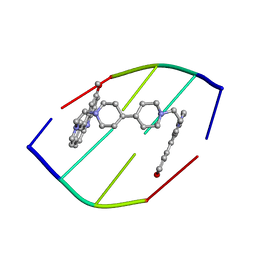 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2O4Q
 
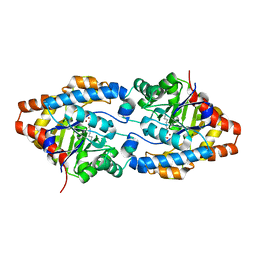 | | Structure of Phosphotriesterase mutant G60A | | Descriptor: | CACODYLATE ION, Parathion hydrolase, ZINC ION | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P.C, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
3CAK
 
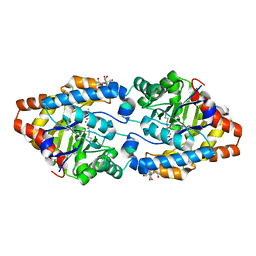 | | X-ray structure of WT PTE with ethyl phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, ... | | Authors: | Kim, J, Tsai, P.-C, Almo, S.C, Raushel, F.M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
3CS2
 
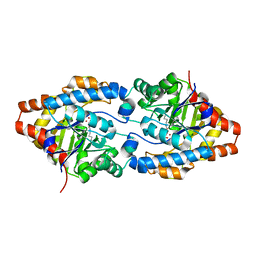 | | Crystal structure of PTE G60A mutant | | Descriptor: | CACODYLATE ION, COBALT (II) ION, Parathion hydrolase | | Authors: | Kim, J, Almo, S.C. | | Deposit date: | 2008-04-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
8DMH
 
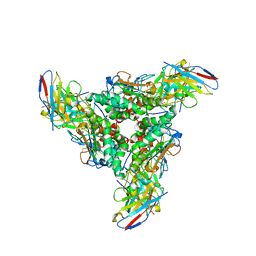 | | Lymphocytic choriomeningitis virus glycoprotein in complex with neutralizing antibody M28 | | Descriptor: | 18.5C-M28 Fab Heavy Chain, 18.5C-M28 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
8DMI
 
 | | Lymphocytic choriomeningitis virus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G1, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
7SKU
 
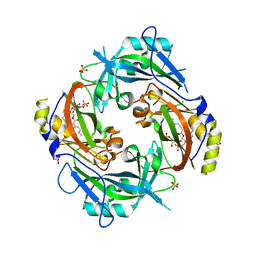 | | Nipah virus matrix protein in complex with PI(4,5)P2 | | Descriptor: | Matrix protein, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Norris, M.J, Saphire, E.O. | | Deposit date: | 2021-10-21 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization.
Sci Adv, 8, 2022
|
|
7SKS
 
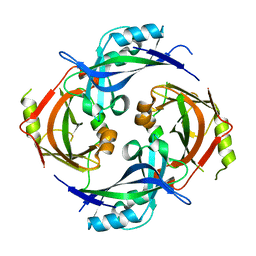 | |
7SKT
 
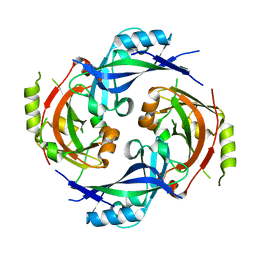 | |
7UOV
 
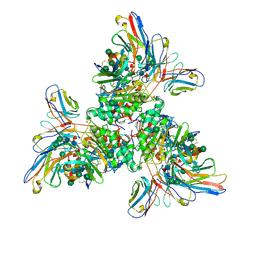 | |
7UOT
 
 | | Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 37.2D heavy chain (variable domain), ... | | Authors: | Li, H, Saphire, E.O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A cocktail of protective antibodies subverts the dense glycan shield of Lassa virus.
Sci Transl Med, 14, 2022
|
|
7UDS
 
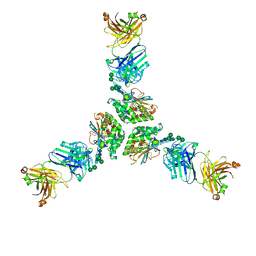 | | Structure of lineage I (Pinneo) Lassa virus glycoprotein bound to Fab 25.10C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25.10C Fab Heavy Chain, ... | | Authors: | Buck, T.K, Enriquez, A.E, Hastie, K.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralizing Antibodies against Lassa Virus Lineage I.
Mbio, 13, 2022
|
|
7UL7
 
 | | Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab | | Descriptor: | 18.5C-M30 Fab Heavy Chain, 18.5C-M30 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Buck, T.K, Enriquez, A.S, Hastie, K.M. | | Deposit date: | 2022-04-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Neutralizing Antibodies against Lassa Virus Lineage I.
Mbio, 13, 2022
|
|
8UST
 
 | | In-virion structure of Ebola virus nucleocapsid-like assemblies from recombinant virus-like particles (nucleoprotein, VP24,VP35,VP40) | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, Polymerase cofactor VP35, ... | | Authors: | Watanabe, R, Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Cell, 187, 2024
|
|
8USN
 
 | | Intracellular cryo-tomography structure of EBOV nucleocapsid at 8.9 Angstrom | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, Polymerase cofactor VP35, ... | | Authors: | Watanabe, R, Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Cell, 187, 2024
|
|
