8R83
 
 | | pentameric IgMFc-AIM complex global refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
8R84
 
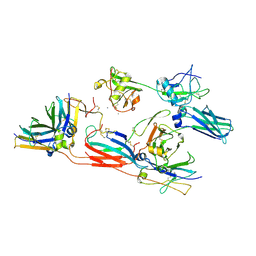 | | pentameric IgMFc-AIM complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CD5 antigen-like, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
8BPG
 
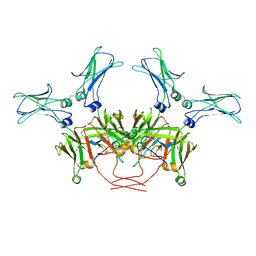 | | FcMR binding at subunit Fcu3 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPF
 
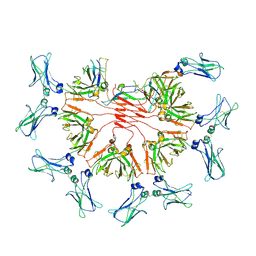 | | FcMR binding at subunit Fcu1 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPE
 
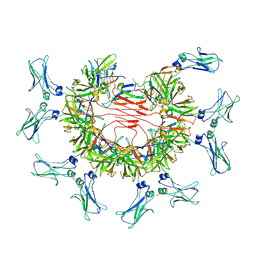 | | 8:1 binding of FcMR on IgM pentameric core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9JUK
 
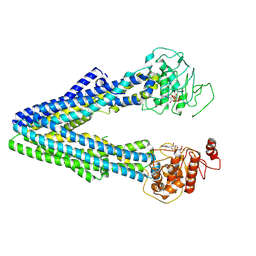 | |
9JUL
 
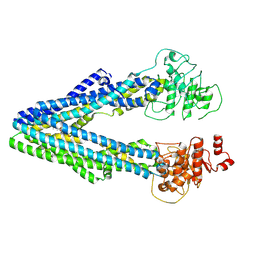 | |
9JUN
 
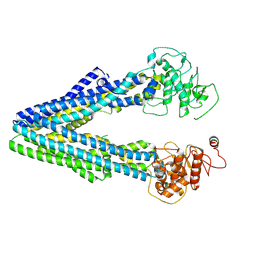 | |
9JUM
 
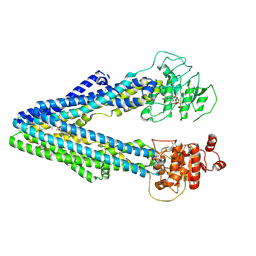 | | Structure of Arabidopsis thaliana ABCB1 with brassinolide and AMP-PNP bound in the inward-facing conformation | | Descriptor: | ABC transporter B family member 1, Brassinolide, MAGNESIUM ION, ... | | Authors: | Chen, Q, Su, N, Guo, J. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures and mechanisms of the ABC transporter ABCB1 from Arabidopsis thaliana.
Structure, 33, 2025
|
|
9JUP
 
 | |
9JUJ
 
 | |
9JUO
 
 | |
7QDO
 
 | | Cryo-EM structure of human monomeric IgM-Fc | | Descriptor: | Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2021-11-27 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
1YFZ
 
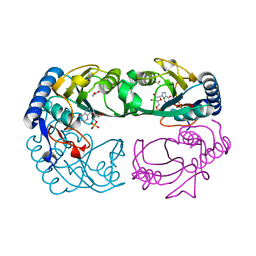 | | Novel IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID, ... | | Authors: | Chen, Q, Liang, Y, Su, X, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
J.Mol.Biol., 348, 2005
|
|
6X67
 
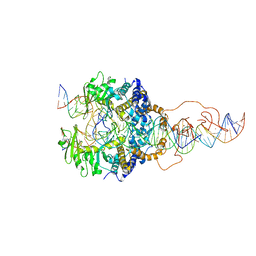 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | Descriptor: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6X68
 
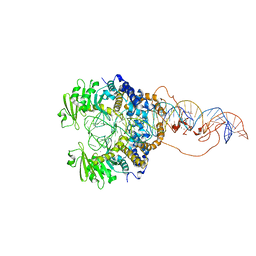 | | Cryo-EM structure of piggyBac transposase synaptic complex with hairpin DNA (SNHP) | | Descriptor: | CALCIUM ION, Transposase, ZINC ION, ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6LDU
 
 | |
6LE7
 
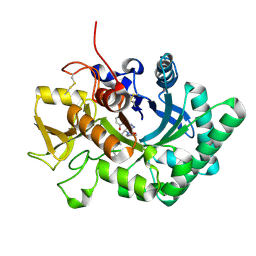 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
6LE8
 
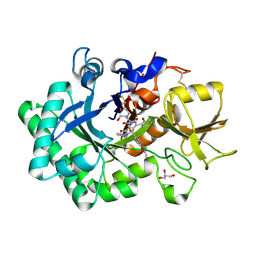 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1 | | Descriptor: | (4R)-4-(4-ethoxyphenyl)-3-(2-hydroxyphenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39909327 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1
To Be Published
|
|
2AIN
 
 | |
7S03
 
 | |
1R3U
 
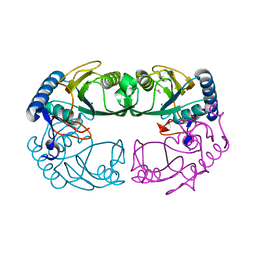 | | Crystal Structure of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Chen, Q, Liang, Y.H, Gu, X.C, Luo, M, Su, X.D. | | Deposit date: | 2003-10-03 | | Release date: | 2004-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis
To be published
|
|
9E82
 
 | |
6J2B
 
 | | CTX-M-64 beta-lactamase S130T sulbactam complex | | Descriptor: | Beta-lactamase, GLYCEROL, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-12-31 | | Release date: | 2019-10-30 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2O
 
 | | Crystal structure of CTX-M-64 clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
