5G2G
 
 | |
4K1U
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y32F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
4K1V
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y57F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
1OV4
 
 | | Crystal structure of human DHEA-ST complexed with androsterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, Alcohol sulfotransferase, SULFATE ION | | Authors: | Chang, H.J, Shi, R, Rhese, P, Lin, S.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex.
J.Biol.Chem., 279, 2004
|
|
5AI1
 
 | | Crystal structure of ketosteroid isomerase containing Y32F, D40N, Y57F and Y119F mutations in the equilenin-bound form | | Descriptor: | EQUILENIN, KETOSTEROID ISOMERASE | | Authors: | Cha, H.J, Jeong, J.H, Kim, Y.G. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Contribution of a Low-Barrier Hydrogen Bond to Catalysis is not Significant in Ketosteroid Isomerase.
Mol.Cells, 38, 2015
|
|
7F92
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein, TETRADECANE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7F93
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in nanodiscs with soybean lipids at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein, TETRADECANE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7F94
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels | | Descriptor: | A C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQF
 
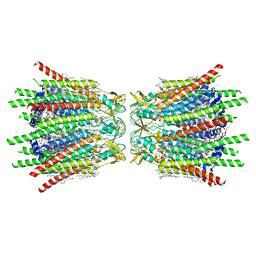 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQD
 
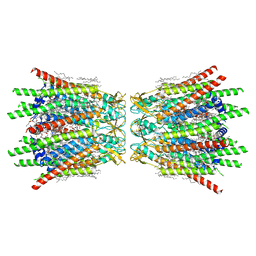 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQG
 
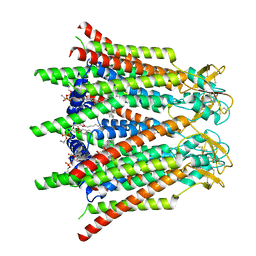 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQH
 
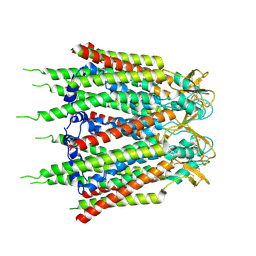 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation) | | Descriptor: | C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQJ
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQI
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQ9
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQB
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
1VZZ
 
 | |
3AUV
 
 | | Predicting Amino Acid Preferences in the Complementarity Determining Regions of an Antibody-Antigen Recognition Interface | | Descriptor: | sc-dsFv derived from the G6-Fab | | Authors: | Yu, C.M, Peng, H.P, Chen, I.C, Lee, Y.C, Chen, J.B, Tsai, K.C, Chen, C.T, Chang, J.Y, Yang, E.W, Hsu, P.C, Jian, J.W, Hsu, H.J, Chang, H.J, Hsu, W.L, Huang, K.F, Ma, A.C, Yang, A.S. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface
Plos One, 7, 2012
|
|
3PJF
 
 | |
3PG4
 
 | |
3PJD
 
 | |
3PJE
 
 | |
4DIT
 
 | |
4UOY
 
 | | Crystal structure of YgjG in complex with Pyridoxal-5'-phosphate | | Descriptor: | FORMIC ACID, GLYCEROL, PUTRESCINE AMINOTRANSFERASE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-06-11 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structure of Putrescine Aminotransferase from Escherichia Coli Provides Insights Into the Substrate Specificity Among Class III Aminotransferases.
Plos One, 9, 2014
|
|
4UOX
 
 | |
