3CWX
 
 | | Crystal structure of cagd from helicobacter pylori pathogenicity island | | Descriptor: | protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
1S2X
 
 | | Crystal structure of Cag-Z from Helicobacter pylori | | Descriptor: | Cag-Z, ISOPROPYL ALCOHOL | | Authors: | Cendron, L, Seydel, A, Angelini, A, Battistutta, R, Zanotti, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system
J.Mol.Biol., 340, 2004
|
|
3CWY
 
 | | Structure of CagD from H. pylori pathogenicity island crystallized in the presence of Cu(II) ions | | Descriptor: | COPPER (II) ION, protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
2G3V
 
 | | Crystal structure of CagS (HP0534, Cag13) from Helicobacter pylori | | Descriptor: | (UNK)(UNK)(UNK)(UNK)(UNK)(MSE)(UNK), CAG pathogenicity island protein 13 | | Authors: | Cendron, L, Tasca, E, Angelini, A, Seydel, A, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CagS from helicobacter pylori
To be Published
|
|
3VC0
 
 | | Crystal structure of Taipoxin beta subunit isoform 1 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino de Laureto, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
3VBZ
 
 | | Crystal structure of Taipoxin beta subunit isoform 2 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
3DJS
 
 | |
3DJZ
 
 | |
3DO4
 
 | |
3DJR
 
 | |
3DJT
 
 | |
3DK0
 
 | |
3DK2
 
 | |
3IWV
 
 | | Crystal structure of Y116T mutant of 5-HYDROXYISOURATE HYDROLASE (TRP) | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
3Q1E
 
 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase in complex with T4 | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Percudani, R, Zanotti, G, Berni, R. | | Deposit date: | 2010-12-17 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
3IWU
 
 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
5LFD
 
 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ramazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-01 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
5LG5
 
 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ragazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
2O73
 
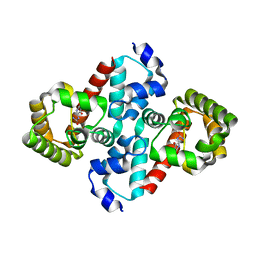 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O70
 
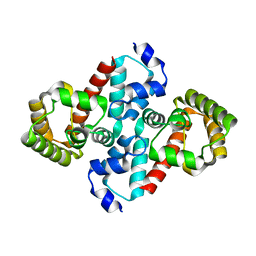 | | Structure of OHCU decarboxylase from zebrafish | | Descriptor: | OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O74
 
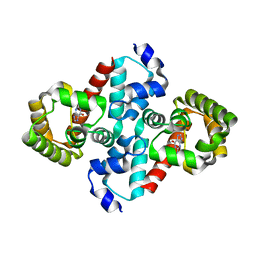 | | Structure of OHCU decarboxylase in complex with guanine | | Descriptor: | GUANINE, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
3KVD
 
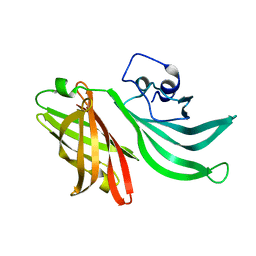 | | Crystal structure of the Neisseria meningitidis Factor H binding protein, fHbp (GNA1870) at 2.0 A resolution | | Descriptor: | Lipoprotein | | Authors: | Cendron, L, Veggi, D, Girardi, E, Zanotti, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the uncomplexed Neisseria meningitidis factor H-binding protein fHbp (rLP2086).
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3QQ5
 
 | | Crystal structure of the [FeFe]-hydrogenase maturation protein HydF | | Descriptor: | Small GTP-binding protein | | Authors: | Cendron, L, Berto, P, D'Adamo, S, Vallese, F, Govoni, C, Posewitz, M.C, Giacometti, G.M, Costantini, P, Zanotti, G. | | Deposit date: | 2011-02-15 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of HydF Scaffold Protein Provides Insights into [FeFe]-Hydrogenase Maturation.
J.Biol.Chem., 286, 2011
|
|
3CXF
 
 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
6Q35
 
 | | Crystal structure of GES-5 beta-lactamase in complex with boronic inhibitor cpd 3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
