6YSY
 
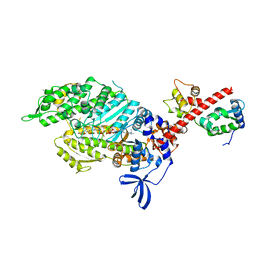 | | Skeletal Myosin bound to MPH-220, MgADP-VO4 | | Descriptor: | (9~{S})-5-methyl-12-(4-morpholin-4-ylphenyl)-9-oxidanyl-4-thia-2,12-diazatricyclo[7.3.0.0^{3,7}]dodeca-1,3(7),5-trien-8-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Canon, L, Kikuti, C.M, Gyimesi, M, Malnasi-Csizmadia, A, Houdusse, A. | | Deposit date: | 2020-04-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Single Residue Variation in Skeletal Muscle Myosin Enables Direct and Selective Drug Targeting for Spasticity and Muscle Stiffness.
Cell, 183, 2020
|
|
2VDE
 
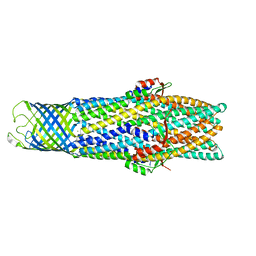 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
2VDD
 
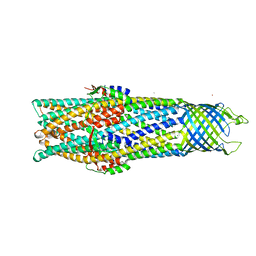 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC, POTASSIUM ION | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
6ER5
 
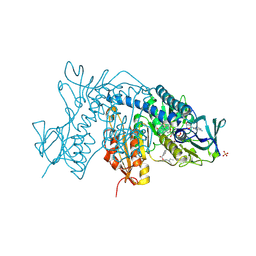 | |
6RB5
 
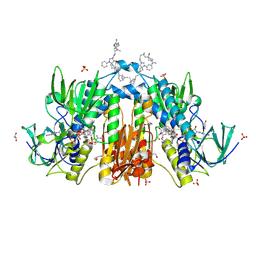 | | Trypanothione reductase in complex with 4-(((3-(8-(2-((1R,2S,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl)ethyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl)propyl)(methyl)amino)methyl)-4-hydroxypiperidine-1-carboximidamide | | Descriptor: | 4-(((3-(8-(2-((1R,2S,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl)ethyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl)propyl)(methyl)amino)methyl)-4-hydroxypiperidine-1-carboximidamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ilari, A, Fiorillo, A, Battista, T, Mocci, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Spiro-containing derivatives show antiparasitic activity against Trypanosoma brucei through inhibition of the trypanothione reductase enzyme.
Plos Negl Trop Dis, 14, 2020
|
|
8ARD
 
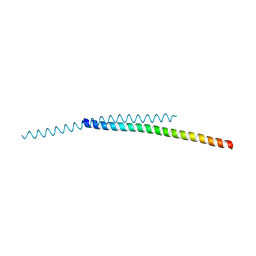 | |
8T2I
 
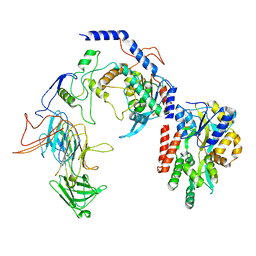 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
6ZP7
 
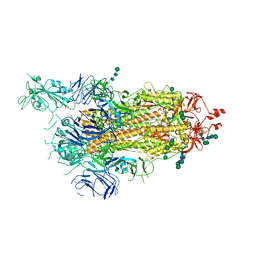 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZOW
 
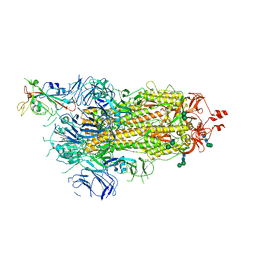 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZP5
 
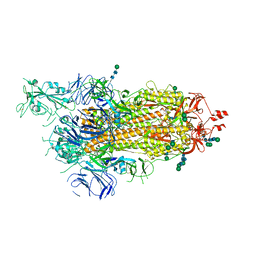 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
