7JJJ
 
 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7JJI
 
 | | Structure of SARS-CoV-2 3Q-2P full-length prefusion spike trimer (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7SBX
 
 | | Structure of OC43 spike in complex with polyclonal Fab6 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBY
 
 | | Structure of OC43 spike in complex with polyclonal Fab7 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB4
 
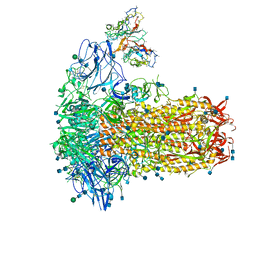 | | Structure of OC43 spike in complex with polyclonal Fab2 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBW
 
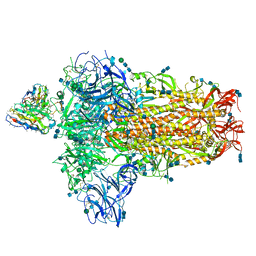 | | Structure of OC43 spike in complex with polyclonal Fab5 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB5
 
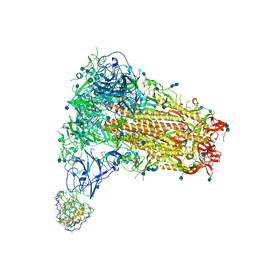 | | Structure of OC43 spike in complex with polyclonal Fab3 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBV
 
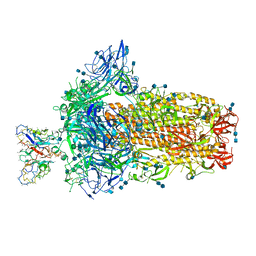 | | Structure of OC43 spike in complex with polyclonal Fab4 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB3
 
 | | Structure of OC43 spike in complex with polyclonal Fab1 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Ward, A, Bangaru, S, Antanasijevic, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
1FAD
 
 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | Descriptor: | PROTEIN (FADD PROTEIN) | | Authors: | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | Deposit date: | 1999-03-23 | | Release date: | 1999-07-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
5W42
 
 | |
6OC3
 
 | |
6OCB
 
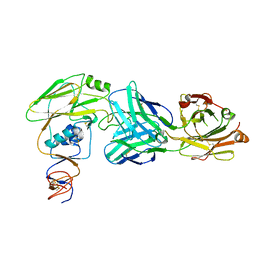 | |
6OBZ
 
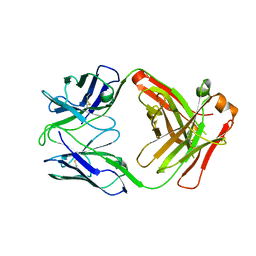 | | Crystal structure of FluA-20 Fab | | Descriptor: | Heavy chain of FluA-20 Fab, Light chain of FluA-20 Fab, Immunoglobulin light chain | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.
Cell, 177, 2019
|
|
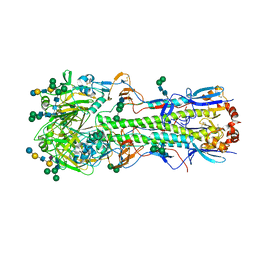
5XRS
 
 | |
5XRT
 
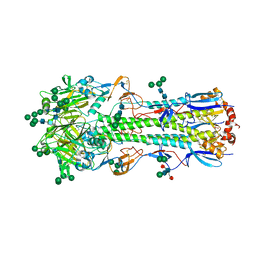 | | Crystal structure of A/Minnesota/11/2010 (H3N2) influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Wilson, I.A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA.
Nat Commun, 9, 2018
|
|
5XRQ
 
 | | Crystal structure of human monoclonal antibody H3v-47 | | Descriptor: | Fab H3v-47 heavy chain, Fab H3v-47 light chain | | Authors: | Zhang, H, Willson, I.A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA.
Nat Commun, 9, 2018
|
|
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
3ELA
 
 | | Crystal structure of active site inhibited coagulation factor VIIA mutant in complex with soluble tissue factor | | Descriptor: | CALCIUM ION, Coagulation factor VIIA heavy chain, Coagulation factor VIIA light chain, ... | | Authors: | Bjelke, J.R, Fodje, M, Svensson, L.A. | | Deposit date: | 2008-09-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of the Ca2+-induced enhancement of the intrinsic factor VIIa activity
J.Biol.Chem., 283, 2008
|
|
8SWH
 
 | |
7JMW
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JMX
 
 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7LM8
 
 | |
7LM9
 
 | | Crystal structure of SARS-CoV spike protein receptor-binding domain in complex with a cross-neutralizing antibody CV38-142 Fab isolated from COVID-19 patient | | Descriptor: | 1,2-ETHANEDIOL, CV38-142 Fab heavy chain, CV38-142 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection.
Cell Host Microbe, 29, 2021
|
|
6PZW
 
 | | CryoEM derived model of NA-22 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-22 fragment antigen binding heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
