8FHC
 
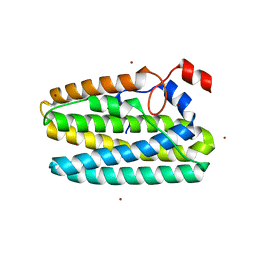 | | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium | | Descriptor: | BROMIDE ION, CHOLIC ACID, FE (III) ION, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium
To Be Published
|
|
8FH5
 
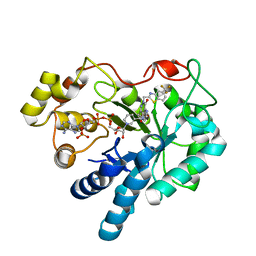 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, Aldo-keto reductase family 1 member B1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001
To Be Published
|
|
8FH9
 
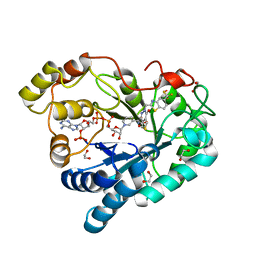 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | Descriptor: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
8FH6
 
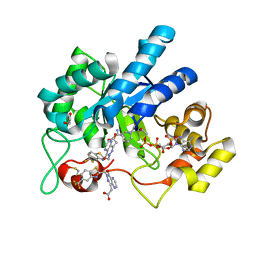 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8FH7
 
 | |
8FHB
 
 | |
8FIF
 
 | |
8FH8
 
 | |
6D9F
 
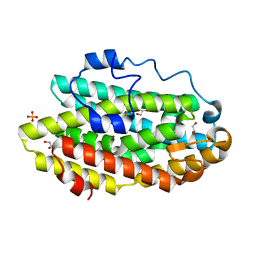 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
5KJK
 
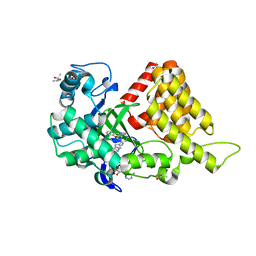 | | SMYD2 in complex with AZ370 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[2-[1-[2-(3,4-dichlorophenyl)ethyl]azetidin-3-yl]oxyphenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJL
 
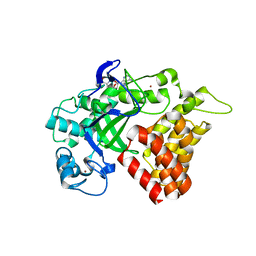 | | SMYD2 in complex with AZ378 | | Descriptor: | N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJN
 
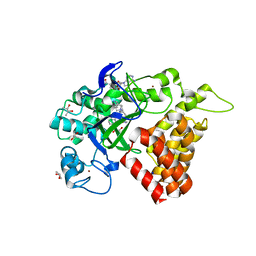 | | SMYD2 in complex with AZ506 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5-[2-[4-[2-(1~{H}-indol-3-yl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-3-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJM
 
 | | SMYD2 in complex with AZ931 | | Descriptor: | 6-[2-[4-[2-(3,4-dichlorophenyl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
3S7B
 
 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
3S7J
 
 | |
3S7D
 
 | |
3S7F
 
 | |
8B6Z
 
 | |
8B6L
 
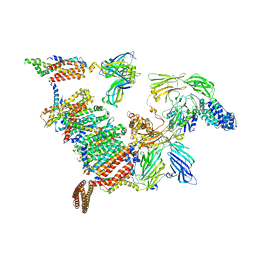 | | Subtomogram average of the human Sec61-TRAP-OSTA-translocon | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2, ... | | Authors: | Gemmer, M, Fedry, J.M.M, Forster, F.G. | | Deposit date: | 2022-09-27 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Visualization of translation and protein biogenesis at the ER membrane.
Nature, 614, 2023
|
|
