5O44
 
 | | Crystal structure of unbranched mixed tri-Ubiquitin chain containing K48 and K63 linkages. | | Descriptor: | MAGNESIUM ION, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2017-05-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | The Crystal Structure and Conformations of an Unbranched Mixed Tri-Ubiquitin Chain Containing K48 and K63 Linkages.
J. Mol. Biol., 429, 2017
|
|
2KHJ
 
 | | NMR structure of the domain 6 of the E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Salah, P, Bisaglia, M, Aliprandi, P, Uzan, M, Sizun, C, Bontems, F. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis
Nucleic Acids Res., 37, 2009
|
|
2KHI
 
 | | NMR structure of the domain 4 of the E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Salah, P, Bisaglia, M, Aliprandi, P, Uzan, M, Sizun, C, Bontems, F. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis
Nucleic Acids Res., 37, 2009
|
|
5IA7
 
 | |
2WLT
 
 | |
2WT4
 
 | |
3F27
 
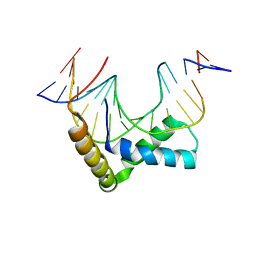 | | Structure of Sox17 Bound to DNA | | Descriptor: | DNA (5'-D(*DCP*DCP*DAP*DGP*DGP*DAP*DCP*DAP*DAP*DTP*DAP*DGP*DAP*DGP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DCP*DTP*DCP*DTP*DAP*DTP*DTP*DGP*DTP*DCP*DCP*DTP*DGP*DG)-3'), Transcription factor SOX-17 | | Authors: | Palasingam, P, Jauch, R, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Sox17 Bound to DNA Reveals a Conserved Bending Topology but Selective Protein Interaction Platforms
J.Mol.Biol., 388, 2009
|
|
3NS7
 
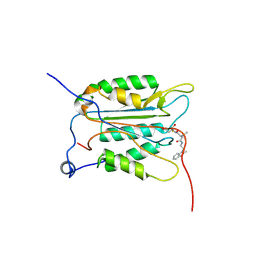 | |
1QBR
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
1QBT
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
8AIJ
 
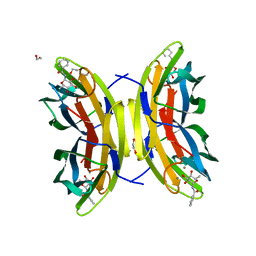 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(alpha-L-Fucopyranosyl)benzamide (6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
8AIY
 
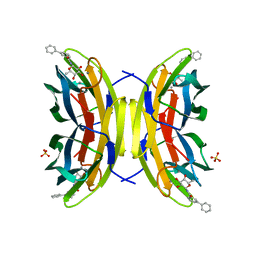 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide (4i) | | Descriptor: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
5IA8
 
 | |
4ICY
 
 | | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kallio, P, Patrikainen, P, Belogurov, G, Mantsala, P, Yang, K, Niemi, J, Metsa-Ketela, M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE.
Biochemistry, 52, 2013
|
|
7NVJ
 
 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NW1
 
 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
2VFI
 
 | | Crystal structure of the Plasmodium falciparum triosephosphate isomerase in the loop closed state with 3-phosphoglycerate bound at the active site and interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1JY4
 
 | |
1JY6
 
 | |
2IPI
 
 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
7ZTC
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 1.6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 1.6, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
7ZTD
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
