5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
8DPM
 
 | |
8DPL
 
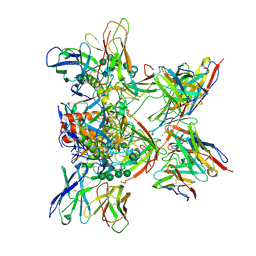 | |
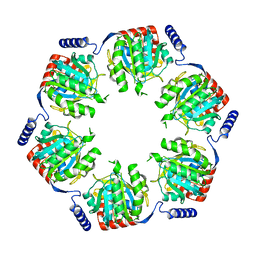
3B8K
 
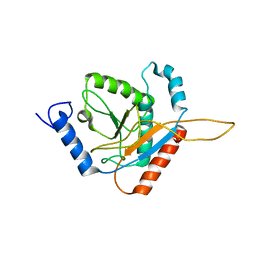 | | Structure of the Truncated Human Dihydrolipoyl Acetyltransferase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase | | Authors: | Yu, X, Hiromasa, Y, Tsen, H, Stoops, J.K, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2007-11-01 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains
Structure, 16, 2008
|
|
2REC
 
 | |
4ZS6
 
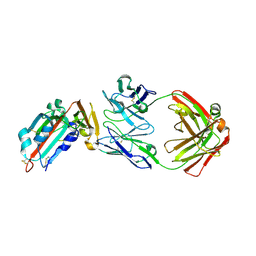 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
3OMH
 
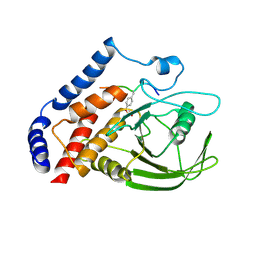 | | Crystal structure of PTPN22 in complex with SKAP-HOM pTyr75 peptide | | Descriptor: | Src kinase-associated phosphoprotein 2, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.-P, Zhang, S, Zhang, Z.-Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2011-09-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate Specificity of Lymphoid-specific Tyrosine Phosphatase (Lyp) and Identification of Src Kinase-associated Protein of 55 kDa Homolog (SKAP-HOM) as a Lyp Substrate.
J.Biol.Chem., 286, 2011
|
|
3FCG
 
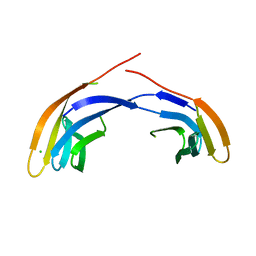 | | Crystal Structure Analysis of the Middle Domain of the Caf1A Usher | | Descriptor: | CHLORIDE ION, F1 capsule-anchoring protein | | Authors: | Yu, X, Visweswaran, G.R, Duck, Z, Marupakula, S, MacIntyre, S, Knight, S, Zavialov, A.V. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Caf1A usher possesses a Caf1 subunit-like domain that is crucial for Caf1 fibre secretion
Biochem.J., 418, 2009
|
|
3CNF
 
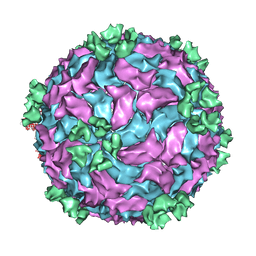 | |
3IZX
 
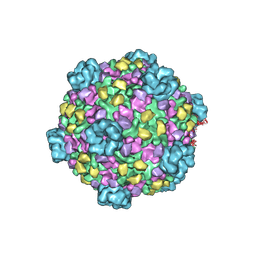 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
3J2V
 
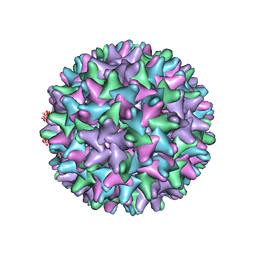 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
3J1R
 
 | | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-terminal Type IV Pilin Helices | | Descriptor: | archaeal adhesion filament core | | Authors: | Yu, X, Goforth, C, Meyer, C, Rachel, R, Wirth, R, Schroeder, G.F, Egelman, E.H. | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-Terminal Type IV Pilin Helices.
J.Mol.Biol., 422, 2012
|
|
3OLR
 
 | |
8A1S
 
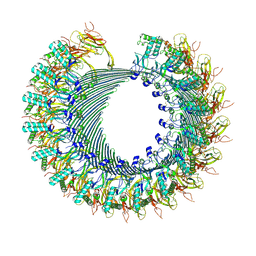 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
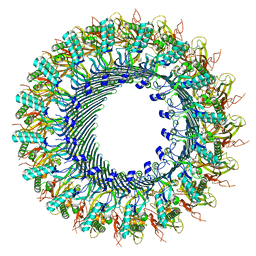 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
5CAZ
 
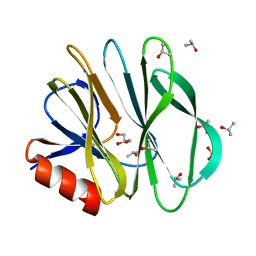 | | Crystallographic structure of apo human rotavirus K8 VP8* | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Outer capsid protein VP4, ... | | Authors: | Yu, X, Blanchard, H. | | Deposit date: | 2015-06-30 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substantial Receptor-induced Structural Rearrangement of Rotavirus VP8*: Potential Implications for Cross-Species Infection.
Chembiochem, 16, 2015
|
|
4BM7
 
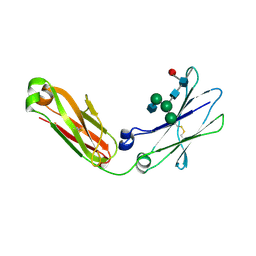 | | Crystal Structure of IgG Fc F241A mutant with native glycosylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, IG GAMMA-1 CHAIN C REGION | | Authors: | Yu, X, Baruah, K, Harvey, D.J, Vasiljevic, S, Alonzi, D.S, Song, B, Higgins, M.K, Bowden, T.A, Crispin, M, Scanlan, C.N. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Engineering Hydrophobic Protein-Carbohydrate Interactions to Fine-Tune Monoclonal Antibodies.
J.Am.Chem.Soc., 135, 2013
|
|
5CB7
 
 | |
5CA6
 
 | |
3TB0
 
 | |
3TAY
 
 | |
6J7O
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7T
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7R
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
8FU7
 
 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
