6BQ1
 
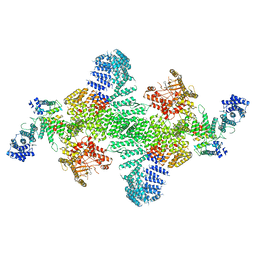 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5K4D
 
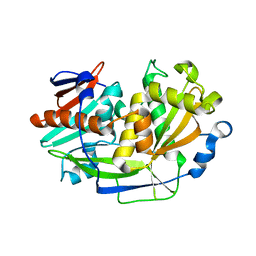 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
6FC4
 
 | |
6FCF
 
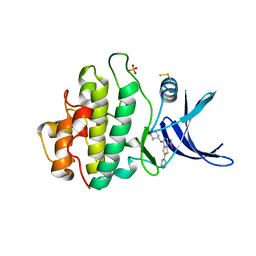 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 44 | | Descriptor: | 4-[[(2~{R},3~{S})-2-methylpiperidin-3-yl]amino]-2-phenyl-thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FCQ
 
 | |
6BTH
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with 2-arachidonoylglycerol (2-AG) | | Descriptor: | 1,3-dihydroxypropan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
5KEE
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature
To be Published
|
|
6CLA
 
 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLJ
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLQ
 
 | | 1.21 A MicroED structure of GSNQNNF at 2.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.21 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CL9
 
 | | 2.20 A MicroED structure of proteinase K at 4.3 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLG
 
 | | 1.35 A MicroED structure of GSNQNNF at 2.4 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.35 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLN
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
5KCE
 
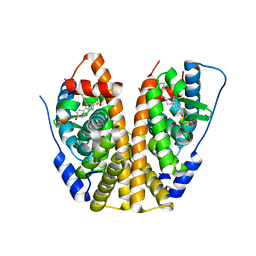 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5JXP
 
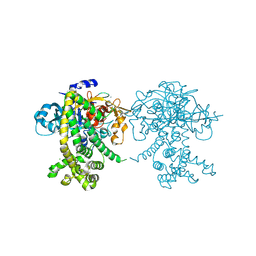 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
6FNP
 
 | | Crystal structure of ECF-CbrT, a cobalamin transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Santos, J.A, Rempel, S, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Functional and structural characterization of an ECF-type ABC transporter for vitamin B12.
Elife, 7, 2018
|
|
1YAI
 
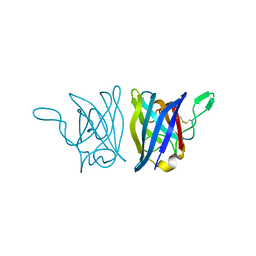 | | X-RAY STRUCTURE OF A BACTERIAL COPPER,ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER, ZINC SUPEROXIDE DISMUTASE, ... | | Authors: | Bourne, Y, Redford, S.M, Lo, T.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 1996-02-03 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel dimeric interface and electrostatic recognition in bacterial Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1YHG
 
 | |
6BYS
 
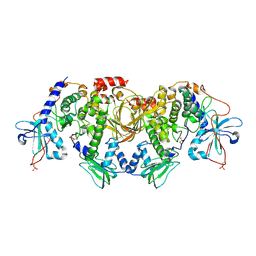 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PRKAc alpha | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
5JH2
 
 | | Crystal structure of the holo form of AKR4C7 from maize | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, Aldose reductase, ... | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
1GUO
 
 | |
6FFI
 
 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
5JCG
 
 | |
2AD5
 
 | | Mechanisms of feedback regulation and drug resistance of CTP synthetases: structure of the E. coli CTPS/CTP complex at 2.8-Angstrom resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of Product Feedback Regulation and Drug Resistance in Cytidine Triphosphate Synthetases from the Structure of a CTP-Inhibited Complex(,).
Biochemistry, 44, 2005
|
|
