1IUJ
 
 | | The structure of TT1380 protein from thermus thermophilus | | Descriptor: | ZINC ION, hypothetical protein TT1380 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TT1380 from Thermus thermophilus HB8
Proteins, 55, 2004
|
|
1J3M
 
 | | Crystal structure of the conserved hypothetical protein TT1751 from Thermus thermophilus HB8 | | Descriptor: | SULFITE ION, the conserved hypothetical protein TT1751 | | Authors: | Kishishita, S, Terada, T, Shirouzu, M, Kuramitsu, S, Park, S.-Y, Tame, R.H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conserved hypothetical protein TT1751 from Thermus thermophilus HB8
Proteins, 57, 2004
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
1IU3
 
 | | CRYSTAL STRUCTURE OF THE E.COLI SEQA PROTEIN COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*TP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-26 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Nucleic Acids Res., 32, 2004
|
|
1IW9
 
 | | Crystal Structure of the M Intermediate of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Takeda, K, Matsui, Y, Kamiya, N, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-25 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the M intermediate of bacteriorhodopsin: allosteric structural changes mediated by sliding movement of a transmembrane helix
J.Mol.Biol., 341, 2004
|
|
1IXE
 
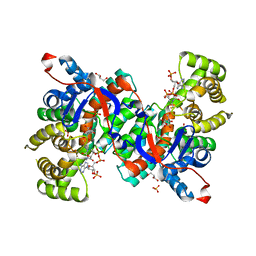 | | Crystal structure of citrate synthase from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Murakami, M, Kanamori, E, Kawaguchi, S, Kuramitsu, S, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural comparison between the open and closed forms of citrate synthase from Thermus thermophilus HB8.
Biophys Physicobio., 12, 2015
|
|
1IV0
 
 | |
1IW6
 
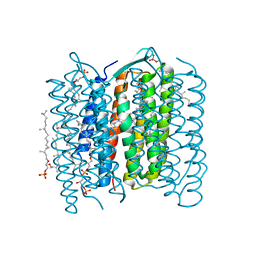 | | Crystal Structure of the Ground State of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Okumura, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary
Photoreaction of Bacteriorhodopsin.
J.Mol.Biol., 324, 2002
|
|
1IXF
 
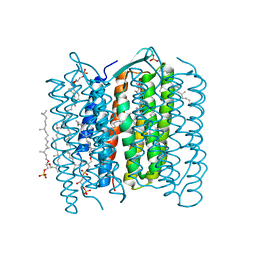 | | Crystal Structure of the K intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Matsui, Y, Sakai, K, Murakami, M, Shiro, Y, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin
J.MOL.BIOL., 324, 2002
|
|
1J3W
 
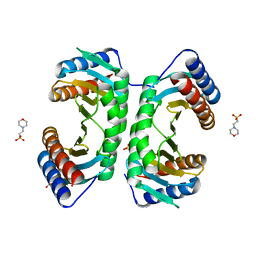 | |
1JZS
 
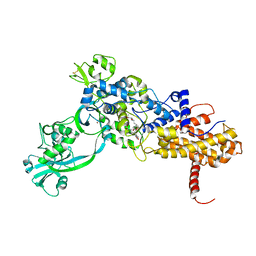 | | Isoleucyl-tRNA synthetase Complexed with mupirocin | | Descriptor: | Isoleucyl-tRNA synthetase, MUPIROCIN, ZINC ION | | Authors: | Nakama, T, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-09-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase.
J.Biol.Chem., 276, 2001
|
|
1IUK
 
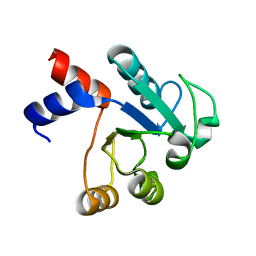 | | The structure of native ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KOY
 
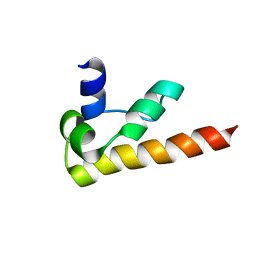 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
1IVZ
 
 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
1J27
 
 | | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7A resolution | | Descriptor: | hypothetical protein TT1725 | | Authors: | Seto, A, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-26 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7 A resolution
Proteins, 53, 2003
|
|
1IVS
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, Valyl-tRNA synthetase, tRNA (Val) | | Authors: | Fukai, S, Nureki, O, Sekine, S.-I, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-29 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
1INZ
 
 | | SOLUTION STRUCTURE OF THE EPSIN N-TERMINAL HOMOLOGY (ENTH) DOMAIN OF HUMAN EPSIN | | Descriptor: | EPS15-INTERACTING PROTEIN(EPSIN) | | Authors: | Koshiba, S, Kigawa, T, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epsin N-terminal homology (ENTH) domain of human epsin
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1J1Y
 
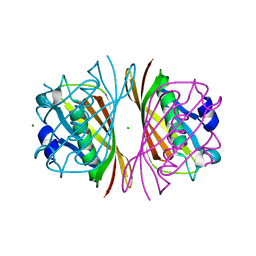 | | Crystal Structure of PaaI from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PaaI protein | | Authors: | Kunishima, N, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1IO7
 
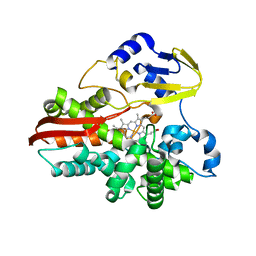 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
1SPK
 
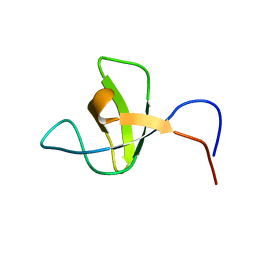 | | Solution Structure of RSGI RUH-010, an SH3 Domain from Mouse cDNA | | Descriptor: | RIKEN cDNA 1300006M19 | | Authors: | Suzuki, Y, Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-17 | | Release date: | 2004-09-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-010, an SH3 Domain from Mouse cDNA
To be Published
|
|
1IT8
 
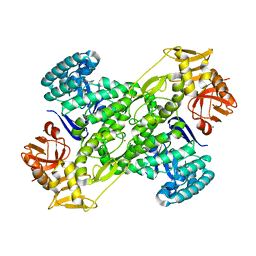 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1J3N
 
 | | Crystal Structure of 3-oxoacyl-(acyl-carrier protein) Synthase II from Thermus thermophilus HB8 | | Descriptor: | 3-oxoacyl-(acyl-carrier protein) synthase II, CITRIC ACID, MAGNESIUM ION | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-oxoacyl-(acyl-carrier protein) synthase II from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1J26
 
 | | Solution structure of a putative peptidyl-tRNA hydrolase domain in a mouse hypothetical protein | | Descriptor: | immature colon carcinoma transcript 1 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality
J.Mol.Biol., 2010
|
|
2YV5
 
 | | Crystal structure of Yjeq from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, YjeQ protein, ... | | Authors: | Wang, H, Kaminishi, T, Hanawa-Suetsugu, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of YjeQ from Aquifex aeolicus
To be Published
|
|
