4X60
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and sinefungin | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
4X61
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAM | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
6WTS
 
 | | CryoEM structure of the C. sordellii lethal toxin TcsL in complex with SEMA6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SEMA6A, ... | | Authors: | Kucharska, I, Rubinstein, J.L, Julien, J.P. | | Deposit date: | 2020-05-03 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of Semaphorin Proteins by P. sordellii Lethal Toxin Reveals Principles of Receptor Specificity in Clostridial Toxins.
Cell, 182, 2020
|
|
4K2F
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4J7R
 
 | |
1Q16
 
 | | Crystal structure of Nitrate Reductase A, NarGHI, from Escherichia coli | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Strynadka, N.C.J. | | Deposit date: | 2003-07-18 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A
Nat.Struct.Biol., 10, 2003
|
|
8BA4
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BA3
 
 | | Crystal structure of JAK2 JH2 in complex with Bemcentinib | | Descriptor: | 1-(3,4-diazatricyclo[9.4.0.0^{2,7}]pentadeca-1(11),2(7),3,5,12,14-hexaen-5-yl)-~{N}3-[(7~{S})-7-pyrrolidin-1-yl-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-3-yl]-1,2,4-triazole-3,5-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8U
 
 | |
8B9H
 
 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B9E
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
6I9Y
 
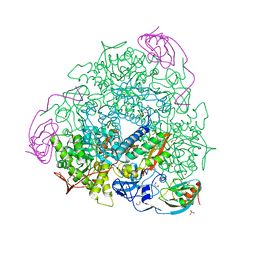 | | The 2.14 A X-ray crystal structure of Sporosarcina pasteurii urease in complex with Au(I) ions | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibition Mechanism of Urease by Au(III) Compounds Unveiled by X-ray Diffraction Analysis.
Acs Med.Chem.Lett., 10, 2019
|
|
5G4H
 
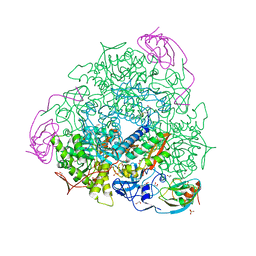 | | 1.50 A resolution catechol (1,2-dihydroxybenzene) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, CATECHOL, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inactivation of Urease by Catechol: Kinetics and Structure.
J.Inorg.Biochem., 166, 2016
|
|
5Y2G
 
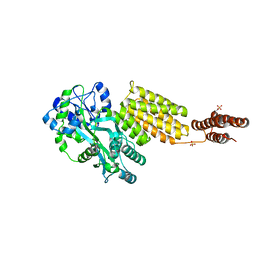 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3ONT
 
 | |
3PDF
 
 | | Discovery of Novel Cyanamide-Based Inhibitors of Cathepsin C | | Descriptor: | 2,5-dibromo-N-{(3R,5S)-1-[(Z)-iminomethyl]-5-methylpyrrolidin-3-yl}benzenesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, B, Laine, D. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of novel cyanamide-based inhibitors of cathepsin C.
Acs Med.Chem.Lett., 2, 2011
|
|
3QNT
 
 | | NPC1L1 (NTD) Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1-like protein 1 | | Authors: | Kwon, H.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Structure of the NPC1L1 N-Terminal Domain in a Closed Conformation.
Plos One, 6, 2011
|
|
2M3K
 
 | |
2O5J
 
 | |
2PPB
 
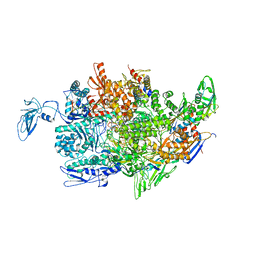 | | Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I, Landick, R. | | Deposit date: | 2007-04-28 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate loading in bacterial RNA polymerase.
Nature, 448, 2007
|
|
