1N5J
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE DIPHOSPHATE (TDP) AND THYMIDINE TRIPHOSPHATE (TTP) AT PH 5.4 (1.85 A RESOLUTION) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5I
 
 | | CRYSTAL STRUCTURE OF INACTIVE MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) AT PH 4.6 (RESOLUTION 1.85 A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRATE ANION, SULFATE ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5K
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE (RESOLUTION 2.1 A) | | Descriptor: | ACETATE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
1N5L
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE, AFTER CATALYSIS IN THE CRYSTAL (2.3 A RESOLUTION) | | Descriptor: | ACETATE ION, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
8EW5
 
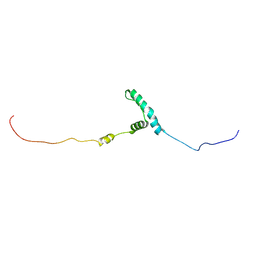 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
6I1I
 
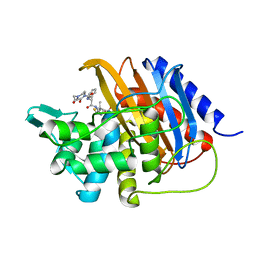 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6I2Z
 
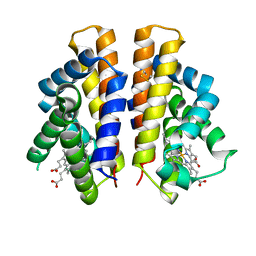 | | Isolated globin domain of the Bordetella pertussis globin-coupled sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Uncharacterized protein | | Authors: | Germani, F, De Schutter, A, Cuypers, B, Berghmans, H, Van Hauwaert, M.-L, Bruno, S, Mozzarelli, A, Moens, L, Van Doorslaer, S, Bolognesi, M, Pesce, A, Dewilde, S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Characterization of the Globin-Coupled Sensors ofAzotobacter vinelandiiandBordetella pertussis.
Antioxid.Redox Signal., 32, 2020
|
|
2CY5
 
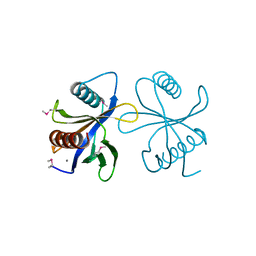 | | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal) | | Descriptor: | CALCIUM ION, epidermal growth factor receptor pathway substrate 8-like protein 1 | | Authors: | Mizohata, E, Hamana, H, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal)
To be Published
|
|
5AHN
 
 | | IMP-bound form of the D199N mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MAGNESIUM ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AHL
 
 | | Apo-form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, SODIUM ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3RWT
 
 | |
2MHS
 
 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
3RWA
 
 | |
5AHM
 
 | | IMP-bound form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, PHOSPHATE ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6N6F
 
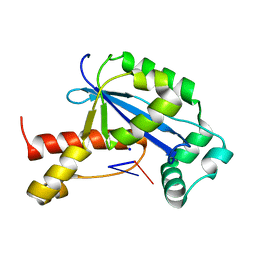 | | Vibrio cholerae Oligoribonuclease bound to pGC | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*GP*C)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
2MZ0
 
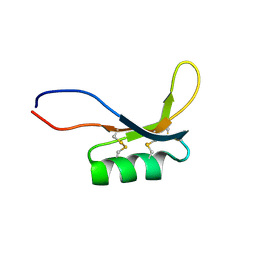 | |
6N6C
 
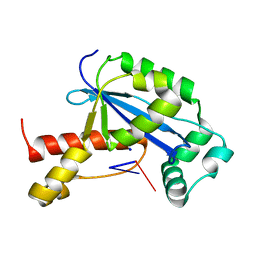 | | Vibrio cholerae Oligoribonuclease bound to pAA | | Descriptor: | RNA (5'-R(P*AP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6J
 
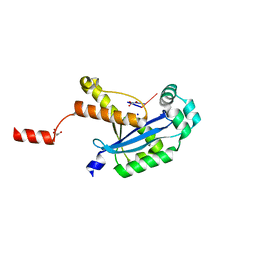 | | Human REXO2 bound to pAA | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MALONATE ION, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6E
 
 | | Vibrio cholerae Oligoribonuclease bound to pGA | | Descriptor: | RNA (5'-R(P*GP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6A
 
 | | Vibrio cholerae Oligoribonuclease bound to pGG | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*GP*G)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6G
 
 | | Vibrio cholerae Oligoribonuclease bound to pCG | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*CP*G)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
3SVO
 
 | |
3SVS
 
 | |
3Q5E
 
 | |
3PZ3
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
