3V06
 
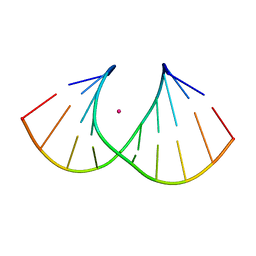 | | Crystal structure of S-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F5H)P*AP*CP*GP*C)-3'), STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
3V07
 
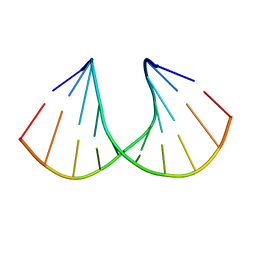 | | Crystal structure of R-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F6H)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
3P4A
 
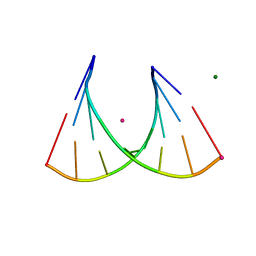 | | 2'Fluoro modified RNA octamer fA2U2 | | Descriptor: | 2'Fluoro modified RNA 8-MER, MAGNESIUM ION, STRONTIUM ION | | Authors: | Manoharan, M, Akinc, A, Pandey, R.K, Qin, J, Hadwiger, P, John, M, Mills, K, Charisse, K, Maier, M.A, Nechev, L, Greene, E.M, Pallan, P.S, Rozners, E, Rajeev, K.G, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
3UKB
 
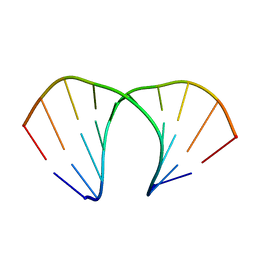 | | (R)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(RCE)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
1XUX
 
 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMS)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
3UKE
 
 | | (S)-cMOE-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(CSM)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3UKC
 
 | | (S)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(1TL)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
1XUW
 
 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMT)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
4HTU
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUF
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*TP*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUE
 
 | | Structure of 5-chlorouracil modified G:U base pair | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(UCL)P*GP*CP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
3P4B
 
 | | Alternatingly modified 2'Fluoro RNA octamer f/rA2U2-P3 | | Descriptor: | 5'-R(*(CFZ)P*GP*(AF2)P*AP*(UFT)P*UP*(CFZ)P*G)-3', BARIUM ION | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
3OPI
 
 | | 7-DEAZA-2'-DEOXYADENOSINE modification in B-FORM DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(7DA)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION | | Authors: | Kowal, E.A, Ganguly, M, Pallan, P.S, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2010-09-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Altering the Electrostatic Potential in the Major Groove: Thermodynamic and Structural Characterization of 7-Deaza-2'-deoxyadenosine:dT Base Pairing in DNA.
J.Phys.Chem.B, 115, 2011
|
|
1EX2
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS MAF PROTEIN | | Descriptor: | PHOSPHATE ION, PROTEIN MAF, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4RBY
 
 | |
4RC0
 
 | |
3P4D
 
 | | Alternatingly modified 2'Fluoro RNA octamer f/rC4G4 | | Descriptor: | 5'-R(*(CFZ)P*CP*(CFZ)P*CP*(GF2)P*GP*(GF2)P*G)-3' | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
3P4C
 
 | | Alternatingly modified 2'Fluoro RNA octamer f/rA2U2-R32 | | Descriptor: | 5'-R(*(CFZ)P*GP*(AF2)P*AP*(UFT)P*UP*(CFZ)P*G)-3', STRONTIUM ION | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
4RBZ
 
 | |
4HUG
 
 | | Structure of 5-chlorouracil modified A:U base pairs | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
1EXC
 
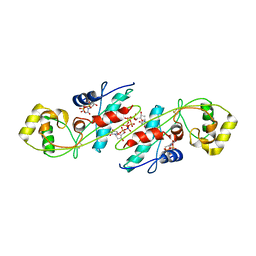 | | CRYSTAL STRUCTURE OF B. SUBTILIS MAF PROTEIN COMPLEXED WITH D-(UTP) | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, PROTEIN MAF, SODIUM ION | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5DO4
 
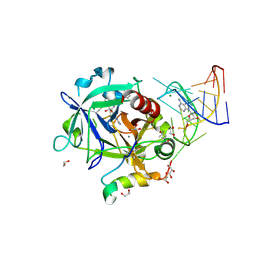 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
4PWM
 
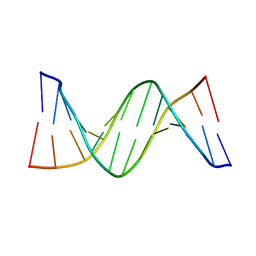 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | Descriptor: | 5'-[CGCGAATT(5CC)GCG]-3' | | Authors: | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
4QC7
 
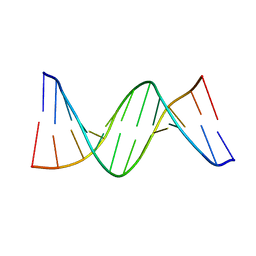 | | Dodecamer structure of 5-formylcytosine containing DNA | | Descriptor: | short DNA strands | | Authors: | Szulik, M.W, Pallan, P, Egli, M, Stone, M.P. | | Deposit date: | 2014-05-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
2AXB
 
 | | Crystal Structure Analysis Of A 2-O-[2-(methoxy)ethyl]-2-thiothymidine Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(S2M)P*AP*CP*GP*C)-3') | | Authors: | Diop-Frimpong, B, Prakash, T.P, Rajeev, K.G, Manoharan, M, Egli, M. | | Deposit date: | 2005-09-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stabilizing contributions of sulfur-modified nucleotides: crystal structure of a DNA duplex with 2'-O-[2-(methoxy)ethyl]-2-thiothymidines.
Nucleic Acids Res., 33, 2005
|
|
