6C6Z
 
 | |
4TLW
 
 | | CARDS TOXIN, FULL-LENGTH | | Descriptor: | ADP-ribosylating toxin CARDS | | Authors: | Becker, A, GALALELDEEN, A, Taylor, A.B, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5V1X
 
 | | Carbon Sulfoxide lyase, Egt2 Y134F in complex with its substrate | | Descriptor: | (1S)-2-{2-[(R)-(2R)-2-amino-2-carboxyethanesulfinyl]-1H-imidazol-4-yl}-1-carboxy-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
4TLV
 
 | | CARDS TOXIN, NICKED | | Descriptor: | ACETATE ION, ADP-ribosylating toxin CARDS, GLYCEROL, ... | | Authors: | Taylor, A.B, Pakhomova, O.N, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CH0
 
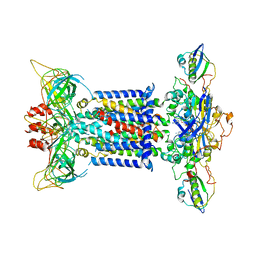 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
4QQC
 
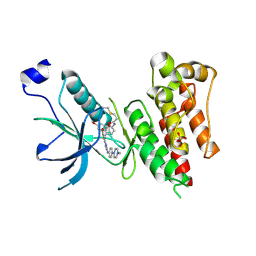 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IZ9
 
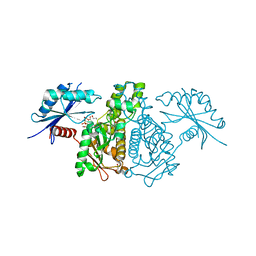 | |
5UTS
 
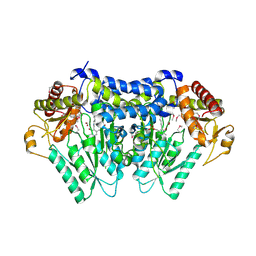 | |
4GK6
 
 | |
4HDT
 
 | |
4Q9Z
 
 | | Human Protein Kinase C Theta in Complex with Compound35 ((1R)-9-(AZETIDIN-3-YLAMINO)-1,8-DIMETHYL-3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | (1R)-9-(azetidin-3-ylamino)-1,8-dimethyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, HUMAN PROTEIN KINASE C THETA, SODIUM ION | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
5V12
 
 | | Crystal structure of Carbon Sulfoxide lyase, Egt2 Y134F with sulfenic acid intermediate | | Descriptor: | (1S)-1-carboxy-2-[2-(hydroxysulfanyl)-1H-imidazol-4-yl]-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
6DI4
 
 | |
6DJ7
 
 | | HIV-1 protease with mutation L76V in complex with GRL-5010 (gem-difluoro-bis-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
4R6V
 
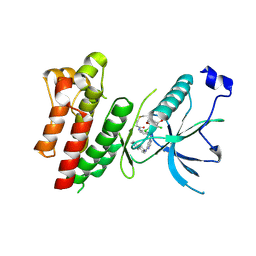 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4I88
 
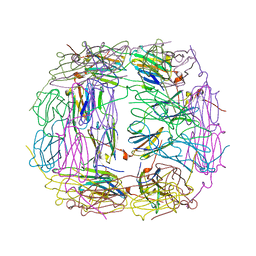 | | R107G HSP16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Pohl, E, Williamson, I.R, Quinlan, R.A. | | Deposit date: | 2012-12-03 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Changes in the quaternary structure and function of MjHSP16.5 attributable to deletion of the IXI motif and introduction of the substitution, R107G, in the alpha-crystallin domain.
PHILOS.TRANS.R.SOC.LOND.B BIOL.SCI., 368, 2013
|
|
4I1Y
 
 | |
4IJN
 
 | |
4EJF
 
 | |
4J5I
 
 | |
8G9S
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC8, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9T
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC9, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAF
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAM
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
